Enzymes Outline - UTCOM Class of 2016

Enzymes
I. Key Properties of Enzymes
A. Enzymes are proteins
1. They can contain non-protein components (cofactors) that participate in catalysis
B. Catalysts
1. Can achieve large enhancements in the rates of reaction
C. Highly Specific
1. Often stereospecific, can distinguish between R and S isomers
D. Ribozymes
1. Small RNAs that act as catalysts
2. Catalytic antibodies also exist
II. Enzyme Basics
A. Enzymes to not change the equilibrium distribution
B. Offer an alternative reaction pathway with a lower activation energy
C. Accelerate reaction rates in both directions
D. Path forward and reverse are the same, just different activation energies
E. Classifications
1. Oxidoreductases – enzymes that catalyze redox reactions
2. Transferases – enzymes that transfer groups to a molecule
3. Hydrolases – enzymes that catalyze hydrolysis reactions
4. Lyases – break C-C or C-N through non-hydrolytic means
5. Isomerases – convert between isomers of the same molecule
6. Ligases – covalently join two reactants with the use of energy (usually ATP)
III. Thermodynamics
A. Gibbs’ Free Energy
1. Independent of reaction path
ΔG°’= -RTlnK eq
= G product
- G reactant
Reactions of biological interest are at pH 7 and 25°C
ΔG rxn
= ΔG°’ + RTlnK eq
ΔG > 0 Reaction is thermodynamically unstable
ΔG = 0 Reaction is at equilibrium
ΔG < 0 Reaction is thermodynamically favorable, “spontaneous”
In this case, the activation energy still must be overcome (E
A
)
2. Coupled reactions a. Energetically unfavorable reactions can be coupled to favorable ones b. Hydrolysis of ATP powers many unfavorable reactions in this way
B. Transition State
1. All chemical reactions have transition states
2. Rate of transition state formation determines rate of the reaction
3. Energy of activation is the energy difference between reactant and TS a. Free energy of TS >> free energy of reactant or product
4. Enzymes provide an alternate pathway through catalysis
C. Catalysis
1. Enzyme forms a complex with substrate
2. Enzyme is not consumed in the reaction
3. Enzyme provides an alternative pathway without altering ΔG of the reaction a. Multiple steps each have lower E
A
4. Enzymes enhance both forward and reverse reactions
D. Activation Energy
ΔG = ΔH – TΔS
Where:
ΔH = change in enthalpy
T = temperature
ΔS = change in entropy (microstates)
ΔH decrease of ΔS increase makes reaction more favorable
IV. Mechanisms of Catalysis
A. Proximity
1. Reactants are brought together, increasing local concentration
2. Enzyme will bind TS better than reactant a. Transition state analogs are good inhibitors
B. Strain and Bond Distortion
1. Substrate binding can induce an active site change that induces strain in substrate
C. Acid-Base a. TS may be induced by strain that increases substrate reactivity
1. Imidazole (histidyl), carboxyl, amino, phenolic, sulfhydryl
2. Protonated forms can act as acid catalysts
3. Deprotonated form can act as base catalysts
4. Apparent pK a
can be affected depending on the group’s access to solvent a. Hydrophobic regions behave more like organic solvent
D. Nucleophilic Catalysis
1. Nucleophilic catalyst can form a covalent complex with a portion of the substrate
RX + Y RY + X
RY + H
2
O ROH + Y + H +
2. Nucleophilic groups include: a. O of Ser-OH – usually unreactive, but environment can increase reactivity b. S of Cys – most reactive protein nucleophile c. N of Lys or His d. COO of Glu and Asp
3. Part of substrate covalently attaches to enzyme and is later released
E. Active Site a. E-S covalent complex is highly reactive
1. Area of enzyme that the substrate binds
2. Reactive groups on the enzyme surface are orientated favorably around reactants
3. Small part of the entire enzyme
4. Often excludes water to provide a nonpolar environment with a lower dielectric a. Makes the electrostatic interactions stronger b. Substrates can be guided by charge distribution c. Charge distribution is also thought to stabilize the TS
5. Noncovalent interactions mediate binding a. Hydrophobics b. Hydrogen bonds c. Ionic interactions d. Van der Waals forces
F. Conformational Flexibility
1. Conformation changes when substrate binds
2. Lock and Key Model (Fischer) a. Older concept that explained specificity (but not catalysis) b. Assumes enzymes are rigid
3. Induced Fit (Koshland) a. Can explain both catalysis and specificity b. Enzymes are flexible and change shape to bind to substrate
E. Ribonuclease A
1. Hydrolyzes RNA after smaller pyrimidine residues (C and U)
2. Requires a 2’OH, so specific for RNA (DNA has no 2’OH)
3. Cleaves the phosphodiester linkage of the backbone
4. Mechanism a. His12 and His119 act as general acid/base catalysts b. His12 deprotonates 2’OH, which nucleophilic attacks phosphodiester i. Forms a cyclic phosphate intermediate i. His119 is deprotonated in the process c. His119 deprotonates H
2
O, which adds in and breaks the ring i. His12 is deprotonated in the process
F. Aspartyl Proteases
1. Asp residue functions in acid/base catalysis
2. Examples include: a. Pepsin b. HIV Protease c. Renin
3. Mechanism a. Aspartyl residue abstracts a proton from H
2
O b. H
2
O nucleophilic adds at C=O center c. Another aspartyl residue protonates the carbonyl following addition d. Aspartyl residue abstracts proton from the former carbonyl i. Electron movement causes peptide bond cleavage e. The first aspartyl residue protonates the nitrogen
G. Serine Proteases
1. Chymotrypsin a. Cleaves at large, hydrophobic residues
2. Trypsin a. Cleaves at arginyl and lysyl residues b. Has an Asp in the active site that stabilizes basic AA binding
3. Elastase a. Cleaves at small residues b. Has Val and Thr side chains in the active site that restrict it sterically
4. Chymotrypsin Mechanism a. Ser195 is activated by deprotonation by His57 (acid/base catalysis) b. Nucleophilic attack by Ser195 on carbonyl of peptide bond i. Addition-elimination mechanism c. His57 protonates the N of the amide d. Oxyanion reforms carbonyl, kicking out the protonated N i. Frees the first part of the peptide e. His57 deprotonates H
2
O, allowing nucleophilic attack on acyl-enzyme i. Addition-elimination by H
2
O f. Reforming carbonyl kicks out the Ser i. Frees second part of the peptide g. Ser-O deprotonates His57, reforming original catalyst structure
H. Prosthetic Groups
1. Tightly bound non-protein component needed for enzyme activity
2. Holoenzyme is the active enzyme a. Apoenzyme is the inactive enzyme b. Holoenzyme = apoenzyme + prosthetic group (cofactor)
3. Vitamins are usually precursors to cofactor substrates used in reactions
4. Examples: a. Heme b. Flavin c. Thiamine d. Metal ions
5. Metalloenzymes a. Enzymes bound to transition metal ions
6. Metal-activated enzymes a. Enzymes that bind metal ions from solution, usually alkali earth metals
7. Function of Metal Ions a. Complexes with substrates (MgATP) b. Mediates redox reactions through changes in oxidation state c. Stabilizes negative charges with electrostatics d. Promotes OH formation at neutral pH
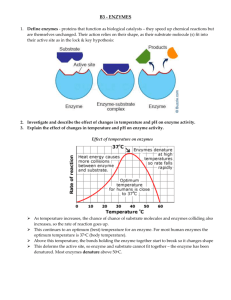
I. Effect of Temperature
1. Activity often increases about 2x per 10°
2. Usually denature at higher temperatures a. Heat denaturation in normally irreversible due to aggregate formation
J. Effect of pH b. Enzymes have an optimal temperature range
1. Activity depends on different groups being ionized
2. Optimal pH range exists, ~7.4 for most enzymes
V. Enzyme Kinetics
A. Michaelis-Menten Kinetics
E + S ES E + P
V
V
0
= (V max max
Where:
[S]) / (K m
+ [S])
= maximum velocity of reaction
S = concentration of substrate
K m
= Michaelis Constant, concentration at which V
0
= ½V max
For our purposes, K m
= K
D
1. Assumptions a. Formation of ES is rapid compared to its conversion to E+P i. We therefore assume that ES is in equilibrium with E+S b. After an initial burst in ES formation, [ES] remains constant c. [S]>>[E], so S in the form of ES is insignificant when considering [S] d. Reaction rate is proportional to [ES] e. The reverse reaction is neglibible
2. Double Reciprocal Equation a. Rearrangement of the Michaelis-Menten Equation yields a linear plot b. Lineweaver-Burke Plot
V
0
= (V max
[S]) / (K m
+ [S]) 1/V
0
= (K m
/V max
)(1/[S]) + (1/V max
)
Y = m
B. Enzyme Inhibitors
1. Competitive Inhibitors a. Usually resemble the substrate b. Inhibitor and substrate bind to the same site c. Effect can be reversed by raising [substrate] d. Do not change V max e. Increase apparent K m
for substrate x + b
K m, apparent
Where:
K
I
= K m
(1 + ([I]/K
= ([E][I])/[EI]
I
)) note that V max
remains constant!
2. Noncompetitive Inhibitors
V a. Usually do not resemble the substrate in question b. Do not prevent binding of substrate in question c. Cannot be overcome by raising [substrate] d. Does not alter K e. Decrease V max, apparent
= V max max
(V m
in unireactant systems max, apparent)
/ (1+ ([I]/K
I
)
C. Turnover Number
1. Number of substrate molecules converted per molecule of enzyme per unit time
2. Turnover number = k cat
3. Varies greatly for different enzymes
4. Can be converted to specific activity
D. Irreversible Inhibitors
1. Enzyme is treated with a reactive reagent that forms a covalent bond with enzyme a. Modified enzyme is inactive b. Inactivation may result from modification of an enzyme in the active site c. Cyanide – modifies heme enzymes by forming a stable heme-complex d. Diisopropylfluorophosphate – reacts with serines in the active site
E. Suicide Inhibitors
1. Specialized substrates that are converted to irreversible inhibitors by the enzyme a. Enzyme proceeds with first step of catalysis then is irreversible inhibited
2. Reactive species formed by enzyme inactivates enzyme
3. Allopurinol – inactivates xanthine oxidase, used to treat gout
VI. Enzyme Regulation
A. Substrate Level
1. Availability of substrate can limit flux through a pathway
B. Allosteric Effectors
1. May activate or inhibit
C. Covalent Modification
1. May activate or inhibit a. Phosphorylation b. Adenylation – bacterial glutamine synthetase c. Methylation – chemotaxis in bacteria
D. Changes in [Enzyme]
1. May result from changes in enzyme synthesis
E. Allosterism and Cooperativity
1. Similar to cooperativity in hemoglobin
2. Allosteric Effectors a. Homoallosterism i. Substrate binding to catalytic site affects neighboring subunits b. Heteroallosterism i. Ligand binds to a distinctly different site from the catalytic site
F. Feedback Inhibition
1. End product inhibits the first committed step in the catalytic pathway
G. Isoenzymes a. Generally through binding to an allosteric site
1. Two or more different enzymes that catalyze the same reaction
2. Result from different gene products
3. May have specific tissue locations
VII. Enzymes and Medicine
A. Therapeutic Targets
1. Methotrexate – dihydrofolate reductase (Cancer chemo)
2. Statin drugs – HMG-CoA reductase (Lowers cholesterol)
3. β-lactam antibiotics – bacterial cell wall inhibition (weakens bacteria)
4. Prilosec – proton pump inhibitor (gastric reflux)
5. Ace Inhibitors – angiotensin converting enzyme (lowers BP)
B. Serum Enzymes in Clinical Diagnosis
1. Varying levels of enzymes in particular conditions
2. Myocardial Infarction a. CPK isoenzymes are found in different levels in MI b. CPK
MB
is a heart specific isozyme that is at much high levels in MI c. Troponin – component of contractile muscle that regulates contraction i. TnT & TnI - isoforms specific for cardiac, skeletal, smooth muscle ii. Used as markers for myocardial infarction d. Myoglobin levels increase immediately following MI e. Based of serum enzyme levels, you can figure out how long since MI i. Levels differ depending on time after MI