Lab 11 - FIU Faculty Websites
advertisement

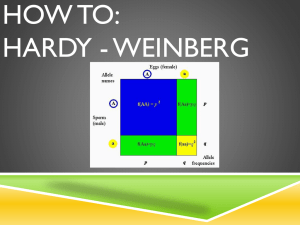
Lab 11: Population Genetics ______________________________________________________________________________ OBJECTIVES: To gain a general understanding about the field of population genetics and how it can be used to study evolution and population dynamics. Understand the concepts of evolution, fitness, natural selection, genetic drift and mutations. To simulate Hardy-Weinberg equilibrium conditions. ______________________________________________________________________________ INTRODUCTION: A population is a group of individuals of one species that occupy a defined geographical area and share genes through interbreeding. Within a large population, new, genetically distinct subpopulations can arise through isolation by distance (IBD). In this mechanism, as the subpopulations become geographically isolated, genetic differentiation between groups in the general population increases. These groups, more commonly referred to as local populations or demes, consist of members that are far likelier to breed with each other than with the remainder of the population. As such, their gene pool differs significantly from that of the general population and with continued isolation, demes may eventually evolve into new species. Demes also arise from other mechanisms including geographical, ecological, temporal and/or behavioral isolation (Fig. 1). Figure 1. Mechanisms of isolation Population genetics is a direct extension of Mendel’s laws of inheritance, Darwin’s ideas of natural selection, and the concepts of molecular genetics. It focuses on the population to which an individual belongs rather than on the individual. Within any given population, every individual has its own set of alleles, and collectively, every individual’s set of alleles comprises the population’s gene pool. The role of a population geneticist is to study the allelic and genotypic (Formulas 1 and 2, respectively) variation present within a population’s gene pool and to assess how this variation changes from one generation to the next. 1. Allelic frequency = # of copies of an allele in a population Total # of all alleles for that gene in a population 1 2. Genotypic Frequency = # of individuals with a particular genotype in a population Total # of all individuals in a population Example: In a population of 100 individuals, 64 are PTC tasters with genotype TT, 32 are PTC tasters with genotype Tt and the last 4 are non-tasters with genotype tt. a. What is the allelic frequency t? Allelic frequency of t = 2*[# inds. w/ recessive genotype (tt)] + # inds. w/ heterozygous genotype (Tt) Total # of individuals in the population = 2[4] + 32 2[100] = 40 200 = 0.2 or 20% b. What is the genotypic frequency of tt? Genotypic frequency of tt = # of tt individuals Total # of individuals in the population = 4 64 + 32 + 4 = 4 100 = 0.04 or 4% In general, populations are dynamic units that change from one generation to the next. To predict how a gene pool changes in response to fluctuations in size, geographic location and/or genetic composition, population geneticists have developed mathematical models that quantify these parameters. The most recognized of these are the Hardy-Weinberg (HW) equations: (p + q)2 = 1 and p2 + 2pq + q2 = 1 These equations relate allele and genotype frequencies in a population and indicates the proportion of each allele combination that should exist within a population. In this formula, p = the frequency of the dominant allele (e.g. B) q = the frequency of the recessive allele (e.g. b) p2 = the frequency of the homozygous dominant genotype (e.g. BB) q2 = the frequency of a recessive genotype (e.g. bb) 2pq = the frequency of a heterozygote genotype (e.g. Bb) 2 The HW equation predicts equilibrium, i.e., the allelic and genotypic frequencies remain constant over the course of many generations, if the following five assumptions are met: (1) Large population size (2) Random mating (3) No mutation (4) No migration (5) No natural selection In reality, no population ever satisfies HW equilibrium completely. Example 1: If in a population of 100 cats, 16 individuals express the recessive phenotype (white fur), then the frequency of the black phenotype is 0.84 and of the white phenotype is 0.16. a. Using the HW equation, calculate the frequencies of alleles B and b. frequency of white (bb) cats = 16/100 = 0.16 q2 = 0.16 therefore q = √0.16 = 0.4 since p + q =1, then p = 1 – q therefore, p =1- 0.4 = 0.6 b. Using the HW equation, calculate the frequencies of the BB and Bb genotypes. From part a, we know that p = 0.6 and q = 0.4 therefore, the frequency of BB cats is p2 = (0.6)2 = 0.36 and the frequency of Bb cats = 2pq = 2(0.6)(0.4) = 0.48 Fig. 11.10 To check: since p2 + 2pq + q2 = 1, then 0.36 + 0.48 +0.16 = 1 3 Example 2: In a population of fruit flies, the genotypes of individuals present are: 50 RR, 20 Rr and 30 rr where R = red eyes and r = white eyes. Assuming the population is in Hardy-Weinberg equilibrium, the proportion of each genotype would be determined as follows: a. Using Formula 1, calculate the frequency of each allele, in this case R and r. Frequency of r allele = 2[30] + 20 2[100] = 80 200 = 0.4 Therefore, q, the frequency of the recessive allele, equals 0.4. b. Since q is known, the p + q = 1 is equation is used to determine p, the frequency of the dominant allele. since p + q =1, then p = 1 – q therefore, p = 1 – 0.4 = 0.6 c. Now using the HW equation, we can calculate the proportion of RR, Rr and rr individuals in the population. From part a and b, we know that p = 0.6 and q = 0.4 therefore, the frequency of RR individuals is p2 = (0.6)2 = 0.36 the frequency of those with the Rr genotype = 2pq = 2(0.6)(0.4) = 0.48 and the frequency of rr flies = q2 = (0.4)2 = 0.16 d. Since there are 100 flies in our population, when the population is in HW equilibirium, 36 flies are homozygous dominant (RR), 48 are heterozygous (Rr) and the remaining16 are homozygous recessive (rr). The genetic composition of a population’s gene pool can be affected by several evolutionary factors, including mutations, migration, non-random mating, genetic drift and natural selection. Mutations, changes in the DNA sequence, are the ultimate source of genetic variation in a population’s gene pool, but because mutation rates are generally low, mutations alone do not usually result in changes in allele frequency. Allelic distributions in a particular group can also fluctuate due to migration, i.e. the movement of individuals either into (immigration) or out of (emigration) a population, resulting in either the addition or loss of alleles, respectively. Diversity within a population is also affected by random events, a process referred to as genetic drift. Two examples of genetic drift are (1) founder effects (Fig. 2A) and (2) genetic bottlenecks (Fig. 2B). In the first scenario, a small group of individuals leave the original population and start a new population in a different location. In contrast, a bottleneck occurs when the original population is drastically reduced in size as a result of some type of 4 natural disaster (e.g. fires, hurricanes, disease, etc.). In addition, non-random mating (e.g. inbreeding/self-fertilization - increases homozygosity) and natural selection also alter the genetic variation within a population. A. B. Figure 2. A. Founder effect and B. Bottleneck effect In summary, the various techniques employed by population geneticists enable them to determine the frequency of gene interaction (gene flow) among different populations and examine the effects of natural selection, migration and mutations, on the genetic composition of these groups. Overall, population genetics attempts to explain how adaptation and speciation shape biological populations over time. In today’s lab you will use the concepts and techniques of population genetics to answer questions about the genetic composition of particular populations. ______________________________________________________________________________ TASK 1 - Genotypic Frequencies and Hardy Weinberg (HW) Equilibrium You can use the HW equations and your knowledge about allelic and genotypic frequencies to address many population genetics questions. The four problems below provide examples. Exercise 1: Gene and Genotype Frequencies In addition to the ABO antigens you learned about in Lab 8, several other classes of glycoproteins are present on the surface of red blood cells. Examples include the MN, Rh, Duffy and Lewis antigens. For this exercise, we will focus on the MN blood protein system. The MN antigens, like the ABO blood proteins, display codominant inheritance where both alleles (i.e., M and N) are expressed simultaneously. In a group of 100 people, 49 are MM, 42 are MN, and 9 are NN. If, M = p and N = q, answer the questions that follow and show all calculations: a. How many M alleles are present among the group? 5 b. How many N alleles are present among the group? c. What is the frequency of the M allele? d. What is the frequency of the N allele? e. What are the frequencies of MM, MN and NN genotypes? Exercise 2: Gene Frequencies in a Medical Application The island of Madagascar has a total population of 1300 people, including 13 individuals who are afflicted by Cystic Fibrosis (CF), a recessively inherited disease. Researchers are interested in knowing how many people in the Madagascar populace are carriers of CF, and have requested your assistance as the resident physical anthropologist. If the C allele is dominant and the c allele is recessive, answer the questions that follow. (Show all calculations) a. What is the frequency of recessive individuals in the Madagascar populace? b. What is the frequency of dominant individuals in this population? c. What is the frequency of the Cc genotype in Madagascar? 6 d. How many people in this population are normal (i.e. not carriers)? e. How many people in this population are carriers? Exercise 3: Examine Your Own Traits Using yourself and your lab mates, complete Tables 1, 2 and 3. Table 1: Individual Observation Representative Trait phenotype Allele Inheritance Your Phenotype Your Character Pattern (check one) possible genotype DOM = dominant, REC = recessive DOM Widow’s Peak W dominant a recessive Darwin’s Ear Point E dominant Cleft Chin c recessive Attached Earlobes 7 REC Ability to detect the bitter taste of phenylthiocarbamide (PTC). Chew on PTC paper to find out your phenotype. Unpigmented Iris (blue) b recessive Tongue Rolling R dominant Tongue Folding f recessive Hitchhiker’s Thumb h recessive Freckles F dominant Mid-Digital Hair M dominant Dimples D dominant PTC tasting T dominant 8 Table 2: Observations for Your Class Trait Total Number Number of Dominant Phenotypes % of Total Number of Recessive Phenotypes % of Total 1. Widow’s Peak 2. Attached Earlobes 3. Darwin’s Ear Point 4. Cleft Chin 5. Unpigmented Iris 6. Tongue Rolling 7. Tongue Folding 8. Hitchhiker’s Thumb 9. Bent Little Finger 10. Mid-Digital Hair 11. Dimples 12. PTC tasting Table 3: Expected Allele and Genotype Frequencies* for Your Class Trait Allele Frequencies p q Genotype Frequencies p2 2pq q2 1. Widow’s Peak 2. Attached Earlobes 3. Darwin’s Ear Point 4. Cleft Chin 5. Unpigmented Iris 6. Tongue Rolling 7. Tongue Folding 8. Hitchhiker’s Thumb 9. Bent Little Finger 10. Mid-Digital Hair 11. Dimples 12. PTC tasting *Note: these are the expected genotypic frequencies if the population is in HW equilibrium. 9 ______________________________________________________________________________ TASK 2 - Beetles Natural Selection Exercise One of the benefits of population genetics and the HW equation is that you can objectively measure the effect of changes in gene frequency due to factors such as natural selection. This concept will be demonstrated using an isolated beetle population with three color variants (red, orange and yellow) in experiments 1, 2 and 3. Note that mating in the beetles is random and only one offspring is produced per mating pair. The colors of offspring follow the pattern below: Mating Pair Red x Red Red x Yellow Red x Orange Orange x Orange Orange x Yellow Yellow x Yellow Offspring Produced Red Orange 50% Red; 50% Orange 50% Orange; 25% Red; 25% Yellow 50% Yellow; 50% Orange Yellow The selection pressure being simulated here is predation. Experiment 1 1. 2. 3. 4. 5. 6. 7. 8. 9. Start the Beetles 2 program by double clicking on the icon on the desktop. Once the program has opened, select New Experiment from the file menu. Ensure that you have a Predator Diet Ratio of 3:3:3. Set a Beetle Population of 30. Set Initial Beetle Numbers to 8:14:8. Model the population by clicking on the ‘10 Generations’ button until Generation 20. Graph your data on the computer and record trend(s). Select "New Experiment" and change the Beetle Population to 60. Repeat the experiment. Select "New Experiment" and change the Beetle Population to 120. Repeat the experiment. Questions: a. Did each experiment show the same trends? b. Which population size showed the greatest variation? Why? c. Which population size was the most stable? Why? 10 d. From these experiments, what can you conclude about genetic variation within a small population as compared to larger ones? e. Is limited diversity an advantage for long term survival of a small isolated population? Explain. Experiment 2: Rates of Predation 1. 2. 3. 4. 5. 6. Select New Experiment from the file menu. Ensure that you have a Predator Diet Ratio of 3:2:1. Set a Beetle Population of 30. Set Initial Beetle Numbers to 10:10:10. Model the population by clicking on the ‘10 Generations’ button until Generation 20. Graph your data on the computer and record trend(s). Questions: a. Why might a predator feed on more red beetles than other colored beetles? b. Why does the number of orange beetles tend toward zero after the red beetle numbers have done so? c. In a natural population, does an individual’s chance of surviving vary from year to year? Explain. 11 Experiment 3: Changing Rates of Predation 1. Select New Experiment from the file menu. 2. Ensure that you have a Predator Diet Ratio of 3:3:3. 3. Set a Beetle Population of 120. 4. Set Initial Beetle Numbers to 30:60:30. 5. Model the population by clicking on the ‘5 Generations’ button. 6. Set the Predator Diet Ratio to 6:1:3. 7. Model the population by clicking on the ‘5 Generations’ button until Generation 10. 8. Set the Predator Diet Ratio to 6:6:1. 9. Model the population by clicking on the ‘5 Generations’ button until Generation 15. 10. Graph your data on the computer and record trend(s). 11. Select "New Experiment" and repeat Steps 2 to 10 three times or combine your data with three other groups. Questions: a. Did increasing the predation rate of red beetles affect the population? Explain. b. Let’s say that red beetles have been selected against. Could they ever completely disappear from the population? c. Can an uncommon variety still be maintained within a population? Explain. 12