Set-up of phytoplankton chemostats in the selection experiment
advertisement

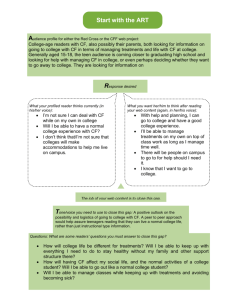
Supporting information Title: Rapid adaptation of herbivore consumers to nutrient limitation: eco-evolutionary feedbacks to population demography and resource control Authors: Steven A.J. Declerck, Andrea R. Malo, Sebastian Diehl, Dennis Waasdorp, Kimberley Lemmen, Konstantinos Proios and Spiros Papakostas Appendix S1 Additional methodological information on the second stage chemostat set-up and the analysis of rotifer cultures Set-up of phytoplankton chemostats in the selection experiment We used phytoplankton chemostats (volume: 1.2 L) with Chlamydomonas reinhardtii as food source for the rotifer populations (Fig. S1). We manipulated phytoplankton stoichiometry by varying the nutrient concentrations in the medium. The medium for the P-replete cultures (HP) consisted of WCmedium with 0.1 mmol L-1 K2HPO4 and 0.48 mmol L-1 NaNO3. The P-limited cultures (LP) received medium composed of WC-medium with 0.02 mmol L-1 K2HPO4 and 2 mmol L-1 NaNO3. Both cultures received 300 mL day-1 of fresh medium and equal light. Figure S1 Photograph of the second stage chemostat system with a phytoplankton chemostat at the top left and four rotifer reactors at the bottom. Collection and culturing of rotifer genotypes prior to selection experiment We incorporated two different pond origins in the design of our experiment to increase the generality of our results. We wanted to increase the robustness of our conclusions by avoiding that our results would be contingent on the genetic architecture of one single population. The distance between the ponds spans approximately 20 km. ‘Pond 7’ (Lat.: 51.854065°, Long.: 5.893175°) is situated in the flood plain of a branch of the lower River Rhine near the city of Nijmegen and has a surface area of about 10 ha. ‘Pond 22’ (Lat.: 51.985263°, Long.: 5.665691°) is located at the campus of Wageningen University, Wageningen and has a surface of approximately 0.25 ha. These two ponds are not hydrologically connected and their B. calyciflorus rotifer populations were found to have moderate levels of genetic differentiation at assumedly neutral microsatellite markers (FST: 0.158; P = 0.001, 10000 permutations; see Tables S2, S3 and S5 for microsatellite data). We refer to Appendix S3 for more details on the development of the microsatellite markers. Resting eggs were separated from pond sediments using a sugar flotation technique (Gómez and Carvalho, 2000) and hatched under light in Petri dishes in distilled water. Upon hatching, B. calyciflorus females were individually transferred into cell culture plates with 2 ml culture medium. Culture medium consisted of C. reinhardtii. The volume of the cultures was progressively increased over the span of one to two weeks to reach 40 ml. Meanwhile, cultures were also divided in two duplicates. Clonal cultures were maintained by transferring each week about half of the culture to a clean tube with 20 ml of fresh culture medium. Set-up of rotifer chemostats in the selection experiment A convenient property of the rotifer reactors in our second stage chemostat system is that they had two ports for medium input, one for food and one for additional medium. Despite the fact that Chlamydomonas concentrations in the phytoplankton chemostats differed among treatments (see above), the two input ports allowed us to provide equal supply rates of food to the rotifer reactors of the different food quality treatments. This was accomplished by blending Chlamydomonas from the first stage chemostats with nutrient-free medium to an incoming food concentration of 1550 µmol C L-1 while keeping dilution rates equal (i.e. 0.19 day-1). We inoculated the rotifer chemostats twice with the same sets of clones, once on the 14/15th of August (day 0 and day 1 of the experiment) and once on the 26/27th of August 2013 (day 12 and day 13). We performed a second inoculation in an effort to reduce initial loss of genetic diversity at low population densities. Clones were represented each by 10 individuals per chemostat during the first inoculation and by 25 individuals during the second inoculation. The dilution medium consisted of WC-medium without N and P. The dilution regime was started on 28 August 2013 (day 14). Soon after, we noticed a systematically lower population density in the wash-out than within the chemostats, indicating avoidance of the chemostat outlets by the rotifers. For this reason, we decided to dilute the rotifer chemostats manually by removing 20% of the medium on a daily basis using a syringe. Rotifer sample analysis Instead of counting rotifer cultures microscopically, we used a FlowCam. A FlowCam is an automatic imaging device (Sieracki et al. 1998) that captures and stores individual images of particles. This allows processing larger volumes of sample per unit of time and effort, higher numbers of particles counted and therefore more reliable density estimates, especially for the rarer objects of interest (e.g. resting eggs and males). We operated our FlowCam with a syringe pump to allow maximum control over sample processing speed and volume. We used a ‘field of view’ (FOV) flow chamber of 300µm depth. Pictures were taken at sample flow rates of 45 ml hour-1 with a frequency of 17 s-1 and an optical magnification of 4. Pictures of particles with a radius above 50µm were all visually inspected, identified and counted by a trained person. Estimates obtained with the FlowCam correlated well with conventional microscopic counts (r=0.83; P<0.0001; n = 71) although we discovered that the FlowCam method resulted in a systematic underestimation of rotifer densities compared to microscopic counts. We accounted for this bias using the regression equation: manual counts = 1.68058 * FlowCam counts. References Gómez, A. & Carvalho, G.R. (2000). Sex, parthenogenesis and genetic structure of rotifers: microsatellite analysis of contemporary and resting egg bank populations. Mol. Ecol., 9, 203–214. Sieracki, C.K., Sieracki, M.E. & Yentsch, C.S. (1998). An imaging in-flow system for automated analysis of marine micro-plankton. Mar. Ecol. Prog. Ser.,168, 285−296. Appendix S2 Stoichiometric seston quality in the phytoplankton and rotifer chemostats of the selection experiment Phytoplankton biomass in the phytoplankton chemostats ranged between 5 and 15 mmol C L-1 in the LP treatment and between 5.6 and 8.5 mmol C L-1 in the HP treatment (Fig. S2). The stoichiometric quality of the phytoplankton food source differed consistently between treatments throughout the course of the selection experiment. Average molar C:P equaled 414 and 61 in the LP and HP chemostats (Fig. S3a), whereas average molar N:P ratios equaled 40 and 4 in the HP chemostat, respectively (Fig. S3c). Dissolved P remained below detection level in both the LP and HP treatments. 20 HP selection LP selection -1 Phytoplankton biomass (mmol L C) The elemental ratios of the seston in the rotifer chemostats differed consistently between treatments and reflected strongly the differences in the phytoplankton chemostats throughout the course of the selection experiment (Fig. S3). Average molar C:P equaled 320 and 72 in the LP and HP chemostats (Fig. S3b), whereas average molar N:P ratios equaled 59 and 11 in the HP chemostats, respectively (Fig. S3d). 15 10 5 0 0 50 100 150 200 250 300 Time (days) Figure S2 Temporal dynamics of the total carbon biomass of C. reinhardtii in the phytoplankton reactors of the second stage chemostat system during the selection experiment. Circles refer to the P-replete chemostat whereas triangles refer to the P-limited chemostat. (a) 800 600 C:P of algae 600 C:P of algae (b) 800 400 400 200 200 0 0 0 50 100 150 200 250 0 300 50 (c) 200 250 300 250 300 (d) 80 60 60 N:P of algae N:P of algae 150 Time (days) Time (days) 80 100 40 20 40 20 0 0 0 50 100 150 200 Time (days) 250 300 0 50 100 150 200 Time (days) Figure S3 (a-d) Temporal dynamics of the elemental composition of C. reinhardtii in the phytoplankton and of seston in the rotifer reactors of the second stage chemostat system during the selection experiment. C:P ratio of C. reinhardtii in the phytoplankton reactor (a), seston C:P ratio in the rotifer reactor (b); N:P ratio of C. reinhardtii in the phytoplankton reactor (c), seston C:P ratio in the rotifer reactor (d). Circles refer to the P-replete chemostat whereas triangles refer to the Plimited chemostat. Due to methodological problems we were unable to obtain reliable data on N during the first 90 days of the experiments. Appendix S3 Microsatellite primer development and application We developed two microsatellite multiplex panels for B. calyciflorus. For this, we used a total of approximately 300 rotifers from a randomly selected clonal line. DNA from these rotifers was extracted with the NucleoSpin® Tissue kit (Macherey-Nagel). Upon testing for DNA quality, construction of the 454 library and 454 GS FLX genome sequencing (Roche Applied Science, Penzburg, Germany) were performed according to the manufacturer’s protocol at the Cytogenetics and Genome Research laboratory of the University of Leuven (Belgium). The 454 run resulted in 473744 DNA sequence reads with an average length of about 393 nucleotides. The raw reads were imported into the QDD v.2.1 pipeline (Meglecz et al. 2010), a tool for microsatellite detection and primer design. We used the software with the default settings, omitting mononucleotide repeats. We selected 24 candidate loci that would give the highest chance of polymorphism (e.g. bi- and trinucleotide repeats) and that would have product sizes and primer sequences that would help generate multiplex panels. Upon testing these candidate loci for successful PCR amplification and polymorphism using rotifers from multiple clonal lines, we developed two multiplex panels of six microsatellite loci each, named SSR1 and SSR2. Microsatellite genotyping We applied the newly developed primers to genotype the clonal lines used to inoculate the rotifer chemostats of the selection experiment. We also applied the same procedure to monitor the chemostat populations during the course of the selection experiment. Rotifer genotyping was performed on DNA extracts from single rotifers using the HotSHOT method (Montero-Pau et al. 2008). In each panel we used, for a 5 μl PCR reaction, 0.5 μl template DNA, 0.1 μl from each primer (from 10 pmol/μl concentration), 2.5 μl Master Mix of the QIAGEN Multiplex PCR kit (2× stock concentration), and 0.8 μl Milli-Q water. PCR thermocycler conditions involved an initial denaturation of 95 oC for 15 min required to activate the HotStarTaq DNA polymerase of the QIAGEN Multiplex PCR kit. The next step was 30 cycles of 30 sec at 95 oC, 90 sec at 56 oC, and 60 sec at 72 oC. A final elongation step of 30 min at 60 oC completed the amplification. The PCR amplicon was then diluted 1:20 with Milli-Q water and 1 μl from this dilution was then mixed with 8.8 μl of formamide and 0.2 μl of GeneScanTM 500 LIZTM size standard (Applied Biosystems, CA) prior loading 1 μl to an ABI Prism 3130 DNA Analyzer (Applied Biosystems, CA). Samples were run for 30 min at 15000 voltage using 36 cm capillaries. Allele calling was performed with the software GeneMapper® v. 4.0 using GS500 (250) LIZ as an option for size standard. Peak threshold was set to 50 rfu for each dye but two different users also did manual inspection and correction, when necessary, of the allele calls. Multilocus genotype (MLG) assignment and analyses of genetic relationship were conducted with GenoDive v.2.b27 (Meirmans & Van Tienderen 2004), a program designed for the analysis of genetic diversity of clonal organisms. To determine the MLG of clonal lines to be inoculated in the chemostats of the selection experiment, three to five rotifers per culture were genotyped. Results for clonal lines originally inoculated in the selection experiment Table S1 shows the nucleotide sequences of the microsatellite primers, the approximate size of the expected PCR products, and the number of alleles observed for the total of clonal lines analyzed. The MLGs of the clonal lines are shown in Tables S2 and S3. Some clonal lines exhibited two different MLGs. For example, in the clonal line BE from ‘Pond 7’, five rotifers were genotyped and two different MLG’s were observed, differing at the genotype of the microsatellite loci A14, 96/96 vs. 96/99, and A15, 87/87 vs. 87/90 (Table S2). Most likely this is the result of selfing as these MLG’s always had the lowest clonal distance and the highest kinship coefficient with each other than with any of the other MLG’s from the same pond (for example, see Table S4). In these cases, both MLGs were kept as representatives of the particular clonal line and they were denoted with different numbers (Tables S2 and S3). During MLG assignment, we omitted information from three loci: (1) locus A3, because of the frequent presence of stutter bands that made genotyping difficult, (2) locus A10, because we found evidence for the presence of null alleles, and, (3) in the case of ‘Pond 22’, from the locus A4, because of too many missing data (Tables S2 and S3). Regardless, the polymorphism present in the rest of the loci was adequate to distinguish each of the clonal lines introduced in the selection experiment. Table S1 Primer sequences, motifs, approximate estimated size (in base pairs, bp) of the PCR product, and number of observed alleles in the clonal lines for the developed microsatellite loci. Locus name Panel Primer sequence (5’ 3’) Motif PCR product (bp) Alleles found (CAA)9 149 5 (AG)7 100 6 (AG)7 161 3 (AC)7 109 3 (TGT)6 96 4 (TAT)6 90 6 (TC)8 125 3 (TGA)8 105 5 (AT)7 181 5 F: AAATGGGCATAAAACGCAAG A1 R: TTCGCGACGAGTTTGTATTG F: AAGCTGTCAACATTGATTTGTTAAA A5 R: CCAAATGTATCCTTCAGGTCA F: AGTTTTGCCTTCAAATTGTATCA A9 R: CTTGTACCCTTGCAATTAACAAC SSR1 F: TTATCTCAACGCCGTTTTGT A11 R: TCAAAACCATTGAATCTAGAAGAA F: TGATATTGAGTTGGCATTGGC A14 R: GCACACCTCAAGCTCAAACA F: AATCGGTACCATTCGTGTCC A15 R: TTGGTTTATTTAAAATCGAGTGATGA F: TGTCTCGTATAGATTAGTTCGACTCC A3 R: CGCTTTAGAAATTGCATTTAGGA F: CGATGTCATTGATGAAGAATCATTT A4 SSR2 R: GAAAATACAGATCAATCAGAATCACC F: CATTGGTAATTCCCGAATTTTG A7 R: TCTTCTTTCCTTAAAGATCAAGTGT F: CGGATTACACAACCATCGAG A8 (AAT)7 238 5 (TTG)7 262 5 (AT)7 145 3 R: TTGGCCTCATCGTCTAAATTG F: TTGTTGGGTTGAATTGACCA A10 R: TGGATGCCATGTTACAAATGA F: TCACGGTCTATTTGGAAACCA A12 R: GGAATTTCGCAAGGCACTTA Table S2 Multilocus genotypes (MLG) for the clonal lines established from the ‘Pond 7’. The size in base pairs (bp) of each allele is separated with “/”. Grey-shaded cells indicate loci that have not been used for clonal assignment of the rotifer samples from the chemostats due to the presence of many stutter bands (=locus A3) or of null alleles (=locus A8). SSR1 SSR2 Culture Name A1 A5 A9 A11 A14 A15 A3 A10 A4 A12 A8 A7 A 143/146 100/100 161/163 109/111 96/99 87/90 121/121 262/268 99/105 147/147 232/235 183/183 H 143/146 96/100 161/163 111/111 96/99 87/90 121/121 262/268 93/105 147/149 229/238 181/181 K 146/146 96/96 163/163 109/109 96/96 87/90 121/121 268/268 99/105 147/147 214/229 181/181 P 146/146 98/98 163/163 109/111 96/99 87/87 121/121 268/268 96/96 147/147 232/232 181/181 S 146/146 98/100 163/165 111/111 96/96 84/90 121/121 262/268 96/105 147/147 235/235 181/181 AD 143/146 98/98 163/163 111/111 99/99 90/90 123/123 262/268 96/96 147/147 238/238 181/181 AE 146/146 96/98 161/163 111/111 96/99 87/87 123/123 262/268 96/105 149/149 238/238 181/183 AG 146/146 96/100 161/163 111/111 96/99 87/90 123/123 268/268 96/102 147/151 238/238 181/187 BD 146/146 96/100 163/163 109/111 96/96 90/93 123/123 262/268 96/105 147/149 235/235 181/183 BE1 143/143 96/96 161/161 109/113 96/99 87/90 123/123 262/268 105/105 149/151 238/238 181/183 BE2 143/143 96/96 161/161 109/113 96/96 87/87 123/123 262/268 105/105 149/151 238/238 181/183 BM 146/146 98/100 161/163 111/111 96/99 87/87 123/123 265/268 105/105 147/149 229/238 181/181 BZ 143/143 96/96 163/163 109/111 96/96 87/96 125/125 262/268 93/105 147/149 229/235 181/187 CW 140/146 96/96 163/163 109/111 96/99 87/90 123/123 Note: For clonal line BE we detected two different MLG’s, possibly as the result of selfing. 262/268 93/93 147/149 235/235 181/183 Table S3 Multilocus genotypes for the clonal lines established from the ‘Pond 22’. The size in base pairs (bp) of each allele is separated with “/”. Grey-shaded cells indicate loci that have not been used for clonal assignment of the rotifer samples from the chemostats due to the presence of many stutter bands (=locus A3) or of null alleles (=locus A8), or of many missing data (= locus A10). SSR1 SSR2 Culture Name A1 A5 A9 A11 A14 A15 A3 A10 A4 A12 A8 A7 E 134/143 96/98 163/163 109/109 96/96 87/93 123/125 259/268 93/93 147/147 232/238 179/185 M 134/143 96/100 161/163 109/111 96/99 90/90 121/125 268/268 96/105 147/149 229/238 183/185 P 143/143 96/96 163/165 109/111 99/99 90/90 125/125 262/271 96/105 147/147 232/238 185/185 Q 143/149 96/100 161/163 109/111 99/99 90/90 123/125 262/268 93/93 147/147 232/238 179/185 U 134/143 96/96 165/165 109/111 99/99 87/90 121/125 262/268 93/93 147/147 232/238 185/187 AD1 134/143 96/98 163/163 109/111 96/99 90/90 123/123 259/268 96/105 147/147 238/238 187/187 AD2 134/143 98/98 163/163 109/111 96/99 90/90 123/123 259/268 96/105 147/147 229/238 185/187 AE 134/143 96/96 161/163 109/111 96/99 81/96 123/125 259/268 96/96 147/149 232/232 181/185 AO 134/143 96/100 163/163 109/111 99/99 90/90 125/125 262/268 93/102 147/147 238/238 185/185 AQ 134/134 96/96 163/163 111/111 99/99 90/90 125/125 259/268 96/96 147/147 232/232 185/185 AZ 143/143 96/98 161/161 109/111 96/99 87/87 121/125 262/262 93/102 147/147 238/238 181/187 BA1 134/143 96/100 161/163 111/111 96/99 87/93 121/125 262/262 96/102 147/149 232/238 181/185 BA2 143/143 96/100 161/163 111/111 96/99 87/93 121/125 262/271 96/102 147/149 232/238 181/185 BC1 134/143 96/96 161/165 109/109 99/99 87/90 125/125 259/268 96/96 147/147 232/238 185/185 BC2 134/143 96/96 161/165 109/109 99/99 87/90 125/125 259/259 96/96 147/147 232/238 185/185 BE 134/143 94/96 161/163 109/109 99/99 90/90 121/125 262/268 96/102 147/147 238/238 185/187 BF 134/143 96/96 161/163 109/109 96/99 90/90 125/125 262/268 93/96 147/147 232/238 185/187 BG1 143/146 98/98 165/165 109/109 96/99 93/93 125/125 262/268 93/93 147/147 232/238 185/185 BG2 143/146 98/100 165/165 109/109 96/99 93/93 123/125 262/268 93/93 147/147 238/238 185/185 BH 146/146 96/96 163/163 109/111 96/96 93/93 123/123 259/262 93/96 147/147 232/238 187/187 BJ1 134/143 96/96 163/165 109/109 96/99 87/90 125/125 262/262 102/105 147/147 232/238 185/185 BJ2 143/143 96/96 163/165 109/109 96/99 87/90 125/125 262/268 102/105 147/147 232/238 185/185 BL 143/143 96/96 161/161 109/109 99/99 81/81 125/125 259/262 93/93 147/147 232/238 185/185 BM1 143/143 96/96 163/163 109/109 96/99 81/81 123/125 262/268 96/102 147/147 232/235 181/181 BM2 143/143 96/96 163/163 109/109 96/99 81/81 125/125 262/268 96/96 147/147 235/235 181/185 BO 134/134 96/96 163/163 109/111 96/99 81/90 125/125 268/268 93/102 147/147 238/238 183/185 BP 143/143 94/98 163/163 109/109 99/99 90/93 125/125 262/262 96/99 147/147 238/238 181/185 BQ 134/143 96/98 163/163 111/111 99/99 90/90 123/123 262/262 93/96 147/147 238/238 179/181 BZ1 146/146 100/100 163/163 109/109 93/99 93/93 125/125 262/271 93/99 147/147 238/238 185/187 BZ2 134/146 100/100 163/163 109/109 93/99 93/93 125/125 262/271 93/93 147/147 238/238 185/187 CA1 143/143 96/100 163/165 109/109 96/96 81/81 121/121 262/268 93/96 147/147 232/238 181/185 CA2 134/143 96/100 163/165 109/109 96/96 81/81 121/121 262/268 93/96 147/147 232/238 181/185 CE 134/146 96/98 165/165 109/109 96/99 87/90 125/125 262/268 93/93 147/147 235/238 181/185 CH 143/143 98/100 163/163 109/109 96/96 81/93 123/125 259/259 93/93 149/149 232/238 179/179 CJ 143/143 96/96 161/161 109/111 96/96 90/90 123/125 268/268 96/96 147/147 238/238 181/181 CK 134/143 96/96 163/163 111/111 90/96 90/90 123/125 259/268 96/102 147/147 238/238 181/185 CM 143/143 96/108 163/165 109/111 96/96 87/90 123/125 262/268 93/102 147/147 235/235 179/181 CN 143/143 96/96 163/163 109/111 96/96 81/90 125/125 262/268 102/102 147/147 238/238 181/181 Note: In each of the clonal lines AD, BA, BC, BG, BJ, BM, BZ and CA we detected two different MLG’s, possibly as the result of selfing. Table S4 Clonal distances and kinship coefficients among the MLG from clonal line BE1 and all other MLG’s from Pond 7. Clonal distance was lowest with the MLG of BE2. Similarly, the kinship coefficient with that MLG was much higher, suggesting that BE1 and BE2 resulted from selfing. BE1 Culture name Clonal distance Kinship coefficient A 42 0.106 H 44 0.243 K 74 0.041 P 67 -0.232 S 55 -0.184 AD 47 0.064 AE 28 0.339 AG 42 0.224 BD 44 -0.043 BE1 - - BE2 6 1.006 BM 39 0.163 BZ 51 0.185 CW 46 -0.088 Table S5 Multilocus genotypes of clones that were additionally genotyped for the assessment of the degree of population genetic differentiation between Pond 7 and Pond 22. Clones were obtained from resting eggs of ‘Pond 7’ and ‘Pond 22’. Fst-values were calculated from these genotypes combined with those of the established clonal lines (see Tables S2 and S3). The size in base pairs (bp) of each allele is separated with “/”. Sample Name SSR1 A1 A5 A9 SSR2 A11 A14 A15 A3 A10 A4 A12 A8 A7 Pond 7 7R 146/146 98/100 163/165 111/111 96/96 84/90 123/123 262/268 96/105 147/147 235/235 181/181 7V 146/146 98/100 163/165 111/111 96/96 84/90 121/121 262/268 96/105 147/147 235/235 181/181 7X 146/146 98/100 163/165 111/111 96/96 84/90 121/121 262/268 96/105 147/147 235/235 181/181 7AA 146/146 98/100 163/165 111/111 96/96 84/90 121/121 262/268 96/105 147/147 235/235 181/181 7AB 146/146 98/100 163/165 111/111 96/96 84/90 123/123 262/268 96/105 147/147 235/235 181/181 7AC 146/146 98/100 163/165 111/111 96/96 84/90 121/121 262/268 96/105 147/147 235/235 181/181 7AH 143/146 96/98 163/165 109/111 96/99 87/87 123/123 268/268 96/105 147/149 232/238 181/185 7AI 143/146 96/98 163/165 109/111 96/99 87/87 121/121 268/268 96/105 147/149 232/238 181/185 7AJ 143/146 96/98 163/165 109/111 96/99 87/87 123/123 268/268 96/105 147/149 232/238 181/185 7AK 143/146 98/98 163/163 109/111 99/99 87/87 123/123 268/268 93/96 147/147 232/238 181/181 7AL 134/146 100/100 163/163 109/109 96/99 90/93 121/121 262/268 93/96 147/149 235/235 181/181 7AM 146/146 100/100 163/163 109/111 99/99 87/90 123/123 268/268 102/105 149/149 229/235 181/185 7AN 134/146 98/98 163/163 109/111 96/96 87/90 123/123 262/262 102/105 147/147 null 179/179 7AP 134/143 96/98 163/165 109/111 93/99 81/87 121/121 262/268 93/99 147/147 238/238 181/181 7AS 146/146 96/96 163/163 111/111 99/99 87/87 123/123 262/268 99/105 147/149 235/235 181/181 7BK 143/146 98/100 163/163 109/109 96/99 81/96 123/123 262/268 93/99 147/147 229/229 181/181 7BN 146/146 98/100 161/163 111/111 96/99 87/87 123/123 265/268 105/105 147/149 229/238 181/181 7BS 134/146 96/96 163/163 111/111 96/99 87/90 123/123 268/268 96/105 147/147 235/235 181/181 7CB 143/146 96/96 161/161 109/111 99/99 87/90 123/123 268/268 96/105 147/149 235/235 181/181 7CQ 134/143 98/98 163/163 109/109 99/99 81/87 125/125 262/268 93/102 147/149 229/229 181/181 Pond 22 22A 134/143 96/96 161/163 109/111 96/96 81/93 121/121 259/268 93/96 147/147 238/238 181/185 22D 143/143 96/96 161/163 111/111 99/99 87/90 125/125 259/268 93/96 147/149 232/232 181/185 22F 134/143 96/96 163/165 109/109 96/99 90/90 123/123 259/268 93/99 147/147 232/232 181/185 22H 134/143 96/98 161/163 109/109 96/99 90/90 123/125 262/268 96/105 147/149 235/235 181/185 22I 134/143 96/98 163/165 109/109 93/96 87/93 123/125 259/268 93/96 147/149 238/238 185/185 22K 134/143 96/100 161/163 111/111 99/99 90/90 123/125 259/268 93/96 147/149 238/238 185/185 22L 134/143 96/102 163/165 109/111 96/96 90/90 123/123 262/268 93/99 147/149 232/232 179/181 22O 134/143 96/98 163/163 109/109 96/99 87/90 125/125 259/268 93/96 147/147 238/238 185/187 22T 134/146 96/98 163/165 109/109 99/99 81/93 121/123 262/268 93/102 147/147 238/238 181/181 22W 134/143 96/96 163/165 109/111 99/99 90/93 123/123 259/268 93/93 147/147 238/238 185/185 22AC 134/134 96/96 161/163 109/109 96/96 90/90 125/125 259/268 93/105 147/149 null 185/187 22AG 134/146 96/96 163/163 109/111 96/96 87/90 123/123 262/268 96/96 149/149 232/232 185/185 22AH 134/146 98/98 165/165 109/109 96/96 87/90 125/125 262/271 105/105 147/147 235/238 179/185 22AJ 143/143 96/96 163/163 109/109 96/96 90/90 121/125 262/262 93/93 147/147 238/238 185/185 22AL 134/134 96/100 161/163 109/109 93/99 90/93 125/125 262/268 93/93 147/147 238/238 185/187 22AP 134/143 100/100 161/163 109/111 96/99 90/90 123/123 259/268 96/102 147/149 232/232 181/185 22AW 134/143 96/96 163/163 109/111 96/99 90/90 121/121 262/268 93/96 147/149 238/238 185/185 22AY 134/143 96/96 163/163 109/111 93/93 90/90 121/125 259/262 93/93 147/147 238/238 181/185 22BD 134/143 96/96 161/161 109/109 96/96 90/90 125/125 262/268 99/105 151/151 null 185/185 22BK 134/143 100/100 161/161 109/109 93/93 90/90 121/121 259/268 96/96 147/147 232/232 181/181 22BS 134/143 96/100 161/163 109/109 96/96 93/93 121/125 262/271 93/93 147/149 238/238 181/183 22BT 134/143 96/96 165/165 109/109 99/99 90/90 121/121 262/268 102/102 147/147 232/238 187/187 22BW 143/143 96/96 163/165 109/111 99/99 90/93 121/121 259/268 93/93 147/147 238/238 185/185 22BX 134/143 98/98 163/165 109/109 96/96 90/90 125/125 262/268 93/102 147/149 232/232 181/187 22BY 134/143 96/100 161/163 109/109 96/99 90/93 123/123 262/268 93/96 147/147 null 181/185 22CB 143/143 94/96 161/163 109/109 99/99 90/90 121/125 262/268 93/96 147/149 null 185/185 22CC 143/146 96/96 163/163 109/109 93/96 93/93 121/121 262/268 93/99 147/147 238/238 185/187 22CD 143/146 96/98 163/163 111/111 96/96 87/90 121/125 262/268 93/105 147/147 238/238 185/187 22CG 143/146 98/100 161/163 109/111 99/99 90/90 121/125 262/268 93/96 147/149 232/238 185/187 22CI 134/143 96/96 163/165 109/111 96/99 90/93 123/125 262/271 99/102 147/147 238/238 185/187 22CL 134/143 96/96 161/163 109/109 96/99 90/90 123/125 262/268 96/96 147/147 232/238 181/185 22CO 143/143 96/100 163/163 109/111 96/99 90/90 125/125 259/268 93/96 147/149 238/238 185/187 References 1.Meglécz, E., Costedoat, C., Dubut, V., Gilles, A., Malausa, T., Pech, N., et al. (2010). QDD: a userfriendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics, 26, 403–404. 2.Meirmans, P.G. & Van Tienderen, P.H. (2004). GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes, 4, 792–794. 3.Montero-Pau, J., Gómez, A. & Muñoz, J. (2008). Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnology and Oceanography-Methods, 6, 218–222. Appendix S4 Summary of linear mixed effects analyses for the selection experiment (see also Fig. 1 in manuscript). Effects of time and the food treatment are presented as the fixed components of the models. Fixed effect SS† MS† d.f. † F† P† Rotifer population density Food 4.69 4.69 15 71.5 <0.001 Time 0.21 0.21 397 7.1 0.008 Food x Time 0.19 0.19 397 6.3 0.013 Food 5.61 5.61 49 63.4 <0.001 Time 0.27 0.27 99 6.2 0.014 Food x Time 0.20 0.20 99 4.5 0.037 Food 3.93 3.93 13 11.7 0.004 Time 67.28 67.28 397 100.9 <0.001 4.21 4.21 397 6.3 0.012 Residual food concentration Per capita number of sexual eggs Food x Time †SS, sum of squares; MS, mean squares; d.f., degrees of freedom; F, F-ratio; P, P-level. Appendix S5 Summary of linear mixed effects analyses for common garden transplant Experiment 2 (see also Figs. 2-4 in manuscript). Effects of the treatments Food, Selection history and Population origin are presented as the fixed components of the models. Fixed effect SS† MS† d.f.† F† P† Rotifer population performance Steady state population biomass Food 1.446 1.446 6 164.0 <0.001 Selection history 0.116 0.116 5 13.2 0.015 Food x Selection history 0.111 0.111 6 12.6 0.012 Population origin 0.067 0.067 5 7.6 0.040 Food 0.689 0.689 6 74.2 <0.001 Selection history 0.068 0.068 5 7.3 0.043 Food x Selection history 0.061 0.061 6 6.6 0.042 Population origin 0.055 0.055 5 5.9 0.060 Food 0.589 0.589 6 326.4 <0.001 Selection history 0.006 0.006 6 3.5 0.109 Food x Selection history 0.055 0.055 6 30.4 0.001 Food 0.235 0.235 38 34.9 <0.001 Selection history 0.028 0.028 6 4.1 0.088 Food x Selection history 0.041 0.041 38 6.1 0.018 Yield Residual seston concentration Rotifer demographic variables Birth (b) and death (d) rates* Per capita number of fecund asexual females Food 3.634 3.634 38 29.1 <0.001 Selection history 0.406 0.406 6 3.3 0.121 Food x Selection history 0.472 0.472 38 3.8 0.059 3.509 3.509 38 9.2 0.004 22.492 22.492 6 58.7 <0.001 6.942 6.942 38 18.1 <0.001 11.142 11.142 38 41.0 <0.001 Selection history 2.594 2.594 6 9.6 0.021 Food x Selection history 2.291 2.291 38 8.4 0.006 0.424 0.424 38 229.5 <0.001 Food 0.087 0.087 38 10.4 0.003 Food 0.896 0.896 38 90.9 <0.001 Food 2.532 2.532 7 1646.2 <0.001 Population origin 0.020 0.020 6 12.8 0.012 Food 2.532 2.532 7 1646.2 <0.001 Population origin 0.020 0.020 6 12.8 0.012 Per capita number of sexual eggs Food Selection history Food x Selection history Per capita number of males Food Rotifer stoichiometry C-content Food P-content C:P Seston stoichiometry Seston C:P Seston N:P *Birth rate b and death rate d yielded identical analysis results †SS, sum of squares; MS, mean squares; d.f., degrees of freedom; F, F-ratio; P, P-level. Degrees of freedom for the food quality effect and its interactions with other factors vary among response variables because replicates for seston stoichiometry and nutrient analyses were pooled before analysis. Appendix S6 Results of common garden transplant experiment 1 Experiment 1 only included treatments with a Pond 22 origin. The results from Experiment 2 that are shown for comparison are therefore also from the treatments with a Pond 22 origin (treatment means across both pond origins are reported in Figs. 2-4). Figure S4 (a-c) Responses of performance related population variables to ambient food quality and selection history in the two common garden transplant experiments. Only treatments with a Pond 22 origin are shown. Steady state rotifer population biomass (a) and yield (b), and residual seston concentration (c). Circles are treatments with a P-replete selection history, triangles are treatments with a P-limitation history. Ambient food quality treatments are: HP: food with high P-content; LP: food with low P-content. Shown are means across chemostat origins ± 2 standard errors (n = 2). Note the log-scale in (a). 300 (a) 0,14 Experiment 1 Experiment 2 Experiment 1 Experiment 2 0,12 0,10 100 90 80 70 60 0,08 Yield Population biomass (µmol C L -1 ) 200 50 0,06 40 30 0,04 HP-selected 20 LP-selected 10 1400 0,02 0,00 HP LP HP LP (c) Experiment 2 1200 1000 800 600 400 200 0 HP HP LP HP Food quality Experiment 1 Residual seston (µmol C L -1 ) (b) LP HP Food quality LP LP Figure S5 (a-d) Responses of demographic rotifer variables to ambient food quality and selection history in the two common garden transplant experiments. Only treatments with a Pond 22 origin are shown. Death rate (a), per capita number of fecund asexual females (b), per capita number of sexual eggs (c) and males (d). Circles are treatments with a P-replete selection history, triangles are treatments with a P-limitation history. Ambient food quality treatments are: HP: food with high Pcontent; LP: food with low P-content. Shown are means across chemostat origins ± 2 standard errors (n = 2). (a) 0,6 Experiment 1 Experiment 2 death rate d (day-1 ) 0,4 0,3 0,2 0,1 HP-selected LP-selected Per capita number of fecund asexual females 0,5 0,0 LP HP Experiment 2 0,5 0,4 0,3 0,2 0,1 HP LP (c) 0,4 Experiment 1 Experiment 2 0,3 0,2 0,1 LP HP LP (d) Experiment 1 Per capita number of males Per capita number of sexual eggs Experiment 1 0,0 HP 0,4 (b) Experiment 2 0,3 0,2 0,1 0,0 0,0 HP LP HP Food quality LP HP LP HP Food quality LP Figure S6 (a-c) Response of rotifer elemental content and elemental ratios to ambient food quality and selection history in the two common garden transplant experiments. Only populations with a Pond 22 origin are shown. Rotifer C-content (a), rotifer P-content (b), molar rotifer C:P ratio (c). Circles are treatments with a P-replete selection history, triangles are treatments with a P-limitation history. Ambient food quality treatments are: HP: food with high P-content; LP: food with low Pcontent. Shown are means across chemostat origins ± 2 standard errors (n = 2). 20 (a) Experiment 1 0,18 Experiment 2 (b) Experiment 1 Experiment 2 0,16 HP-selected Rotifer P content (nmol ind. -1 ) Rotifer C content (nmol ind. -1 ) LP-selected 15 10 5 0,14 0,12 0,10 0,08 0,06 0,04 0,02 0 0,00 HP 200 LP (c) Experiment 1 HP LP Experiment 2 HP-selected LP-selected Rotifer C:P 150 100 50 0 HP LP HP LP HP LP HP LP