Figure Legends for Supplemental Figures Supplemental Figure S1
advertisement

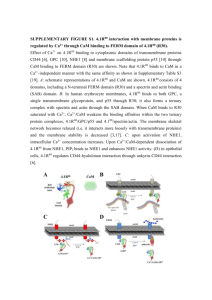
Figure Legends for Supplemental Figures Supplemental Figure S1. Ribbon structure model for 4.1G FERM domain and mapping of binding sites for various binding partners. The model was constructed in silico based on the known 3D structure of the FERM domains of 4.1R (IGG3) and 4.1B (2HE7) [16]. The previously published Ca2+-independent CaM binding site [15] and band 3 binding motif LEEDY [2,18] in 4.1R are both shown as ball-stick models. Note that the LEEDY motif in 4.1R is replaced by a LEKDY motif in 4.1G and that the lysine residue (K) does not affect 4.1G binding to band 3 [16]. Other displayed binding sites include binding sites for GPC, p55 and CD44 which have been all previously characterized [17-19]. Supplemental Figure S2. Zimm plot analysis for GHP and the GHP/Ca2+/ CaM complex. The values for Kc/I(0) were evaluated from the intercepts of the Guinier plots shown in Figure 2 and plotted as a function of protein concentration. The plots for both samples followed a straight line over the entire concentration range measured. The intercept and slope of the lines enabled us to calculate the molecular weight (Mw) and the second virial coefficient (A2), respectively, as described in Equation 4 (see Experimental section). Plots of Kc/I(q,c) ratios at the dilution limit I(0,c) (upper panel) and of the square of the radius of gyration, Rg2 (Å2) (bottom panel) as a function of protein concentration, c (g/cm−3), for GHP (open circles) and GHP/Ca2+/CaM complex (closed circles). Supplemental Figure S3. Titration experiment for Ca2+-bound CaM interaction with with pepG. The spectra at [pepG]/[Ca2+/CaM] ratios of 0.7 1.4 and 2.1 were generated. Spectra at 1.4 and 2.1 ratios were almost identical, suggesting that the binding between Ca2+-bound CaM and pepG occured at a 1:1 stoichiometric ratio. Supplemental Figure S4. NMR spectra of 15N-labelled apo-CaM in absence or presence of pepG. 1 H-15N HSQC spectra for apo-CaM alone and for the pepG/apo-CaM complex. All samples were adjusted at pH 6.8 and kept at 25°C during the experiments. Circles and dotted circles highlight apo-CaM signals that either appear or disappear after addition of pepG, respectively. Highlighted amino acids exhibited a change in NMR spectrum upon pepG binding to apo-CaM. Blue and red dots refer to apo-CaM alone and mixtures at ratio of [pepG]/[apo-CaM]=2.0, [CaM]=0.7mM, [KCl]=100mM, [EDTA]=10mM, pH 6.3, 25°C, respectively. Supplemental Figure S5. NMR spectra of 15N-labelled apo-CaM in absence or presence of pepG. 1 H-15N HSQC spectra for the pepG/apo-CaM complex. All samples were adjusted at pH 6.8 and kept at 25°C during the experiments. Circles and dotted circles highlight apo-CaM signals that either appear or disappear after addition of pepG, respectively. Highlighted amino acids exhibited a change in NMR spectrum upon pepG binding to apo-CaM. The blue, red and orange dots refer to apo-CaM, and to mixtures at ratio of [pepG]/[apo-CaM]=2.0, and [pepG]/[apo-CaM]=3.5, [CaM]=0.7mM, [KCl]=100mM, [EDTA]=10mM, pH 6.3, 25°C, respectively. Supplemental Figure S6. Zimm’s plot analysis for pepG and the pepG/Ca2+/CaM complex. The values of Kc/I(0), evaluated from the intercepts of the Guinier plots shown in Figure 6, were plotted as a function of protein concentration. The plots for both samples followed a straight line over the entire concentration range measured. The intercept and slope of the line enabled us to calculate the molecular weight (Mw) and the second virial coefficient (A2), respectively, as described in Equation 4 (see Experimental section). Supplemental Figure S7. Proposed Models for Ca2+/CaM-dependent regulation of 4.1G binding to membrane proteins. Two models for Ca2+/CaM-dependent regulation of 4.1G binding to membrane proteins are presented. In the first model, GHP keeps its extended shape and forms a complex with Ca2+/CaM. Binding of Ca2+/CaM to the intrinsically disordered GHP induces profound conformational changes in 4.1G FERM domain that can no longer bind to band 3, p55 or CD44. In the second model, as discussed in Figure 9B, the intrinsically disordered GHP bends while forming a complex with Ca2+/CaM (GHP/Ca2+/CaM). The resulting complex wraps the FERM domain, and physically inhibits its binding to band 3 and p55 [16].