Supplementary Material A Decadentate Gd(III)
advertisement

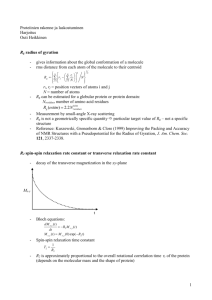
Supplementary Material A Decadentate Gd(III)-Coordinating Paramagnetic Cosolvent for Protein Relaxation Enhancement Measurement Xin-Hua Gu1,3 • Zhou Gong1,3 • Da-Chuan Guo1 • Wei-Ping Zhang2,* • Chun Tang1,* 1 State Key Laboratory of Magnetic Resonance and Atomic Molecular Physics and Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan, Hubei Province 430071, China; 2 Department of Pharmacology, Zhejiang University School of Medicine, Hangzhou, Zhejiang Province, China; 3 X. Gu and Z Gong contributed equally. * e-mail: weiping601@zju.edu.cn (W.-P. Zhang) or tanglab@wipm.ac.cn (C.Tang) Fig. S1 Water transferred PRE using Gd(III)-DTPA-BMA as paramagnetic cosolvent observed for maltose-binding protein (MBP, bound with β-cyclodextrin; PDB accession code 1DMB). The data were extracted from reference (Madl et al. 2009). The residues that experience PRE values much larger than predicted values include 5, 18, 42, 45, 53-54, 92, 101, 124, 128, 143, 174-175, 207, 246, 252-253, 272, 327, 352-356 and 370, with their amide protons shown as red spheres. The two perspectives are related by an 180º rotation. Fig. S2 sPRE measured for protein GB1 using Gd(III)-DTPA-BMA cosolvent. Observed PRE values are shown as red spheres; calculated sPRE values, based on PDB structure 2GB1, are shown as a black line. Error bar stands for 1 standard deviation. Residues broadened out beyond detection are denoted as asterisks. Inset, amide protons of residues with large PRE discrepancies indicated on the protein structure: orange balls, larger observed sPRE values than calculated; red balls, residues that disappeared upon adding paramagnetic cosolvent. Fig. S3 Structure models for Gd(III)-DTPA-BMA and Gd(III)-TTHA-TMA. Gd(III)-TTHA-TMA appears more like a sphere than Gd(III)-DTPA-BMA does. Approximately, the radii for Gd(III)-DTPA-BMA and Gd(III)-TTHA-TMA are 3.5Å and 4.0Å, respectively. The starting structures for the two models are based on references (Ehnebom and Pedersen 1992; Burdinski et al. 2009). Fig. S4. sPRE values for protein GB1 with addition of two different concentrations of Gd(III)-TTHA-TMA. Asterisks denote residues that are too broad to be properly resolved. Lines connect the residues simply for guiding the eyes. Error bars denote 1 standard deviation. Fig. S5. Comparison between the crystal and NMR structures available for protein GB1. In the crystal structure (1PGB), the carboxylate group of Asp22 forms a hydrogen bond with the hydroxyl group of Thr25, and side chain of Asp22 sits on the top of the amide groups of Ala23 and Ala24. In the NMR structure (2GB1), the hydrogen bond is likely absent, as the side chain of Asp22 swings away. Thus, the amide groups of Ala23 and Ala24 are more exposed. Fig. S6 sPRE measurement for MBP bound with maltotriose, upon addition of 5mM Gd(III)-TTHA-TMA paramagnetic cosovlent. No residues are broadened out beyond detection or experiencing PREs larger than 70s -1. The error bars denote 1 standard deviation. Lines are simply used to guide the eyes. References Burdinski, D, Pikkemaat, JA, Lub, J, de Peinder, P, Nieto Garrido, L, Weyhermuller, T (2009) Lanthanide complexes of triethylenetetramine tetra-, penta-, and hexaacetamide ligands as paramagnetic chemical exchange-dependent saturation transfer contrast agents for magnetic resonance imaging: nona- versus decadentate coordination. Inorg Chem 48: 6692-6712 Ehnebom, L, Pedersen, BF (1992) Molecular and Crystal-Structure of a Lanthanide Complex, Dydtpa-Bma Hydrate. Acta Chem Scand 46: 126-130 Madl, T, Bermel, W, Zangger, K (2009) Use of relaxation enhancements in a paramagnetic environment for the structure determination of proteins using NMR spectroscopy. Angew Chem Int Ed Engl 48: 8259-8262