Proteiinien rakenne ja laskostuminen
advertisement
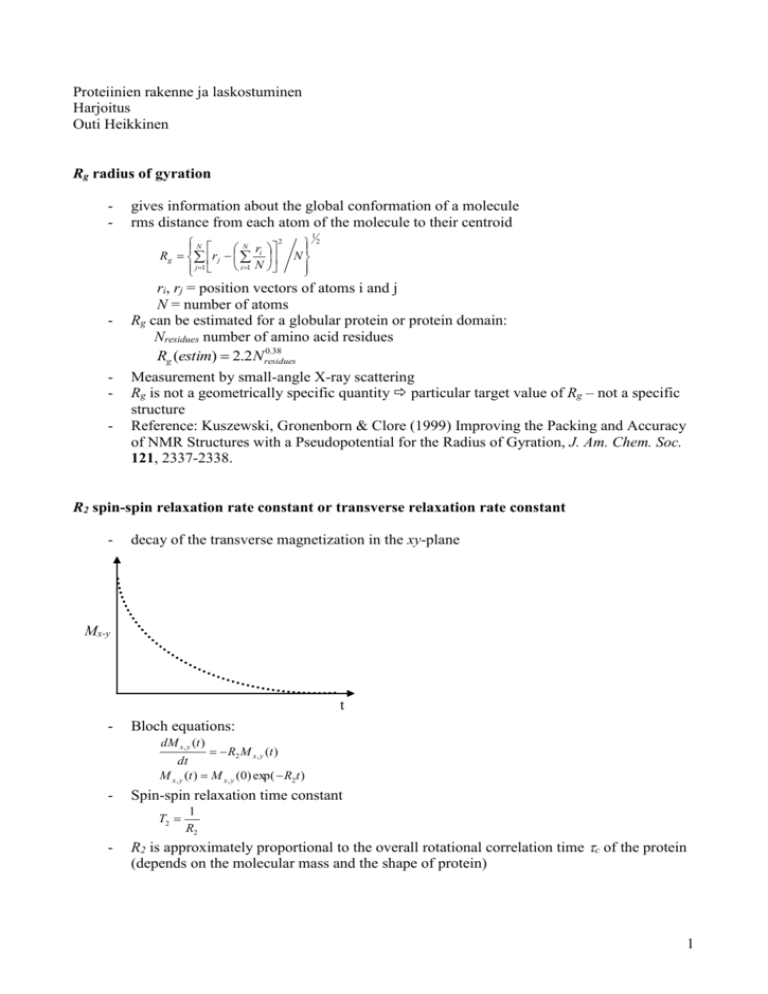
Proteiinien rakenne ja laskostuminen Harjoitus Outi Heikkinen Rg radius of gyration - gives information about the global conformation of a molecule rms distance from each atom of the molecule to their centroid 2 N N ri R g r j N i 1 N j 1 - - 1 2 ri, rj = position vectors of atoms i and j N = number of atoms Rg can be estimated for a globular protein or protein domain: Nresidues number of amino acid residues 0.38 Rg (estim) 2.2 Nresidues Measurement by small-angle X-ray scattering Rg is not a geometrically specific quantity particular target value of Rg – not a specific structure Reference: Kuszewski, Gronenborn & Clore (1999) Improving the Packing and Accuracy of NMR Structures with a Pseudopotential for the Radius of Gyration, J. Am. Chem. Soc. 121, 2337-2338. R2 spin-spin relaxation rate constant or transverse relaxation rate constant - decay of the transverse magnetization in the xy-plane Mx-y t - Bloch equations: dM x , y (t ) R2 M x , y (t ) dt M x , y (t ) M x , y (0) exp( R2t ) - Spin-spin relaxation time constant T2 - 1 R2 R2 is approximately proportional to the overall rotational correlation time c of the protein (depends on the molecular mass and the shape of protein) 1 J ( ) c 1 ( c ) 2 R2 4 J (0) 3J ( N ) ... - (homogenous) linewidth ∆υFWHH in an NMR experiment: ∆υFWHH = R2/π (Hz) FWHH = full-width at half height Spin label - paramagnetic label incorporated into the protein structure o chelated metal ions (heme group) o stable organic radicals e.g nitroxides attached to reactive amino acid side chains Protein S S Cysteine residue N O - induces a distance dependent paramagnetic relaxation enhancement effect on nuclei nearby K r sp R2 - 3 c 4 c 1 h2 c2 1 6 r = the distance between the nuclear spin and the electron spin τc = the correlation time for the electron-nuclear interaction ωh = the Larmor frequency of the nuclear spin R2sp = the paramagnetic relaxation enhancement R2sp = R2* - R2 R2* = the transverse relaxation rate constant for paramagnetic species R2 = the transverse relaxation rate constant for diamagnetic species K = 1/15·S(S+1)γ2g2β2 S = the electron spin γ = the nuclear gyromagnetic ratio g = the electronic g factor β = the Bohr magneton provides long-range distance, tens of angstroms, restraints for protein structure determination by NMR in contrast to short, few angstroms, inter proton distances Reference: Battiste & Wagner (2000) Biochemistry 39, 5355-5365 2