An ontology-based GIS for genomic data management of rumen
advertisement
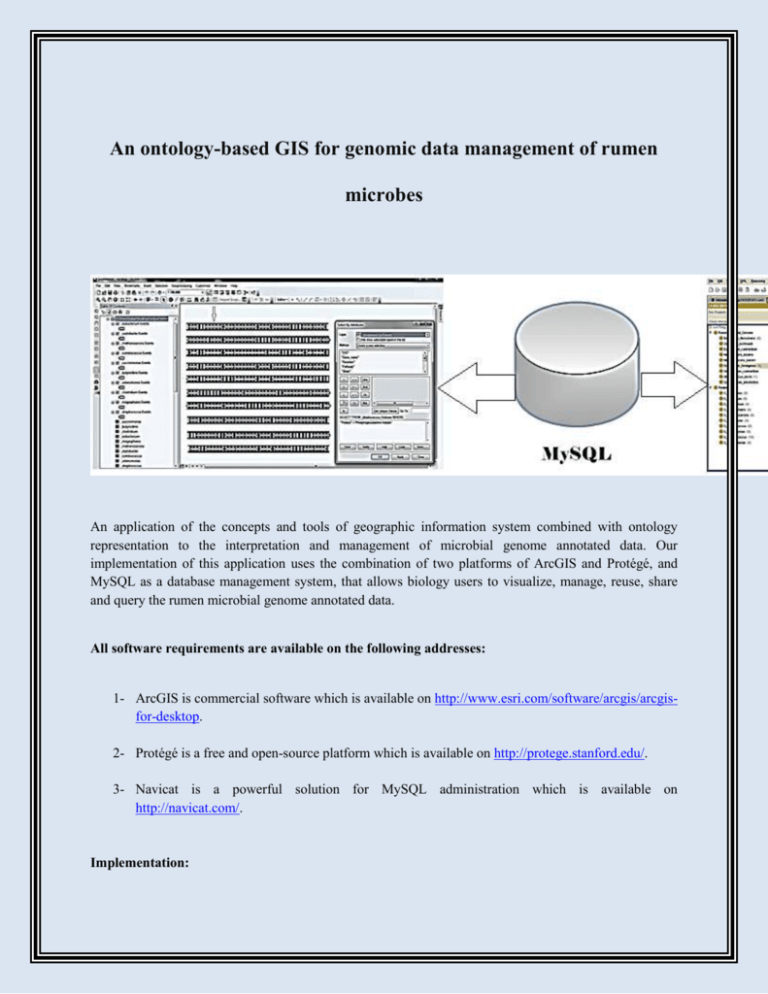
An ontology-based GIS for genomic data management of rumen microbes An application of the concepts and tools of geographic information system combined with ontology representation to the interpretation and management of microbial genome annotated data. Our implementation of this application uses the combination of two platforms of ArcGIS and Protégé, and MySQL as a database management system, that allows biology users to visualize, manage, reuse, share and query the rumen microbial genome annotated data. All software requirements are available on the following addresses: 1- ArcGIS is commercial software which is available on http://www.esri.com/software/arcgis/arcgisfor-desktop. 2- Protégé is a free and open-source platform which is available on http://protege.stanford.edu/. 3- Navicat is a powerful solution for MySQL administration which is available on http://navicat.com/. Implementation: 1- Download the ArcGIS (free trial), Protégé (free) and Navicat (free trial) 2- Install ArcGIS and then softwares Protégé and Navicat 3- Get a sample excel file containing genome annotated data http://patricbrc.org/portal/portal/patric/Home. from PATRIC: The application was developed in two steps. In the first step, the genome annotated data ( e.g., Locus tag, Product name, Ec Number) was extracted from excel file and then prepared and stored in MySQL database. The second step involved the connection of database to both ArcGIS and Protégé as GIS engine and ontology platform, respectively. This work was conducted as part of a PhD thesis at Ferdowsi University of Mashhad and the National Institute of Genetic Engineering and Biotechnology, Bioinformatics Lab.