Detecting Animal Contaminates in Food
advertisement

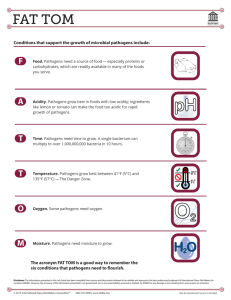
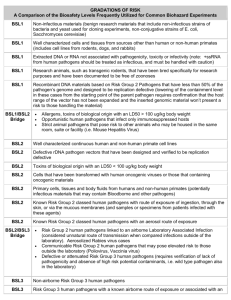
Detecting Animal Contaminates in Food Introduction Food safety is a global health goal and the foodborne diseases take a major crisis on health. Food borne illness has increased over the past few years and although there are an enormous number of different tests for the pathogens that are found in food, there is no one test that can be utilized. The vital problem is unhealthy food from animal contaminates that result in pathogens that are ingested. If one test can be found that can accurately identify multiple pathogens (the most prevalent species in food) using an efficient cost effective technology platform. What technology can be designed that will rapidly detect and characterize the contamination of animal-based food? There are numerous technologies for the detection of several types of pathogens, each with its own limitations. The seeker wishes to find the most efficient and promising technology that will detect the largest variety of pathogens rapidly using a cost effective platform. Conventional and standard bacterial detection methods such as culture and colony counting methods, immunology-based methods and polymerase chain reaction based methods, may take up to several hours or even a few days to yield an answer. This is inadequate, and recently many researchers are focusing towards the progress of rapid methods. Although new technologies like biosensors show potential approaches, further research and development is essential. There are new biomolecular techniques for food pathogen detection that are being developed to improve the biosensor characteristics such as sensitivity and selectivity, also which are rapid, reliable, effective and suitable for in situ analysis. Background on tests Conventional methods for microbiological tests have been based on culturing the microoganisms found to contaminate food. This was time consuming and laborious, requiring culturing the bacteria and producing media. This also requires trained personnel. Nucleic acid based methods such as PCR, multi-plex PCR and quantitative PCR require trained personnel and specialized instruments, however, the tests are highly specialized and specific for detecting certain pathogens. Nucleic acid sequence-based amplification (NASBA) based kits are specific and low cost but require viable micro-organisms for the test. Loop-mediated isothermal amplification (LAMP) is a novel nucleic amplification method that is also very sensitive but the sequence has to be known, thus it is limited in the species it can detect. DNA microarrays are commercially available but are designed for gene expression analysis studies and are expensive. Optical biosensors are not completely developed for detecting pathogens, there are still some problems with their use although they are specific, and are also expensive. ELISAs are highly specific but require trained personnel and specialized equipment. The development of new and improved tests are needed to help prevent the increase in food borne illness by detecting the pathogens in food. Thus, the microbiological analysis of food is limited and remains a challenging task for virtually all assays and technologies developing kits for detection , especially for detecting particular pathogenic species. The problems may be due to : • The complexity of food matrices and composition. • The heterogeneous distribution of low levels of pathogens. • The stress suffered by the microorganisms during the processing of foods. • The presence of bacteria from the normal microbiota, especially in raw foods. The complexity of food matrices remains the major obstacle to the development of effective sampling and rapid testing methods as well as the low amount of pathogen present in the food and large number of species. Long duration enrichments are often used due to the low number of pathogenic microorganisms that tend to be present in food samples. The test needs to have a very low detection limit of a high number of species (both viable or non-viable), thus it must be highly specific, fast, and easy to perform (not needing someone with highly technical skills). The modification and development of some of the present tests may be enough or a new technology all together may be needed. References V. Velusamy, K Arshak, O Korostynska, K Oliwa, C Adley. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnology Advances, Vol. 28: Iss 2 , pp 232-254. Jodi Woan-FeiLaw, Nurul-SyakimaAb Mutalib, Kok-GanChan and Learn-HanLee Rapidmethodsforthedetectionoffoodbornebacterialpathogens:principles,applications,advantag esandlimitations Frontiers in Microbiology The Challenge The seeker specifically wishes to identify a robust method that: 1. 2. 3. 4. 5. Minimizes the time for detection to hours. Easy to use and cost effective and can be done without highly specialized training. Freedom to practice: no IP, however licensing opportunities will be considered. Can detect both viable and nonviable pathogens. Does not rely on the sequencing of the pathogens it detect to be available. In Scope 1. Protocols that describe assays that include procedural steps with reagents and instruments. 2. Scientific explanations and reasons for the recommended techniques. 3. Reference to papers or clinical trials supporting the techniques. Out of Scope The Seeker is not interested in a solution that: • Consists of a literature survey • Requires excessively expensive reagents or supplies • Has many or time-consuming steps • Is primarily theoretical and would be difficult to assess efficacy in a practical manner • Has negative health and safety issues or environmental impact Criteria that must be met: 1. Level of detail provided for the proposed protocol such as reagents, solution formulations, consumables, and equipment. 2. Applicability to high throughput, or a high number of samples 3. Accessibility and cost of the technological solution, i.e., does not require development of an entirely new technology. 4. Ease of method for use at the point of care Project Deliverables Challenge solutions should provide: 1. A detailed description and explanation of the proposed solution or solutions; 2. Persuasive scientific rationale and, ideally, experimental data, to justify the solution; and, 3. Full disclosure of any third party intellectual property used in the solution. \