Electronic Supplementary Material Reactive oxygen metabolites
advertisement

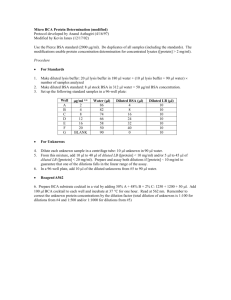
Electronic Supplementary Material Reactive oxygen metabolites (ROMs) and Total plasma antioxidant capacity (TAC) : We measured reactive oxygen metabolites (ROMs) using the d-ROMs test (Diacron International, Grosseto, Italy), which measures the level of hydroperoxides, compounds that signal lipid and protein oxidative damage. We diluted 10 l of plasma in 200 l of the provided acidic buffer solution, and then read the plate kinetically (one read per minute) for the next 10 minutes. Absorbance was measured at 490nm (BioTek ELx800, VT, USA) and we calculated change in ROMs concentrations (in mM of H2O2 equivalents) from these absorbencies by taking the difference between the reading at minute 20 and minute 0, dividing by 20 minutes, and multiplying by the constant 9000 (all per the manufacturers specifications). All samples were run in a single assay plate. The total plasma antioxidant capacity (TAC) was measured using the OXYAdsorbent test (Diacron International, Grosseto, Italy), which measures the effectiveness of the blood antioxidant barrier by quantifying its ability to cope with oxidant action of hypochlorous acid (HClO). We diluted 10 l plasma in 990 l of distilled water; we then mixed 5 l of this diluted plasma with 195 l of the provided HClO solution and continued by following the manufacture’s instructions. We measured absorbance at 490nm (BioTek ELx800) and we calculated TAC (in mM of HClO neutralized). All samples were run in a single assay plate. Single Cell Gel Electrophoresis (SCGE) assay: The SCGE assay uses fresh blood samples and because only a limited number of samples can be run at one time we only explored measures of DNA damage in the IW and IS group. Leukocytes were diluted in 1X phosphate buffered saline, suspended in 0.75% low melting point agarose, and plated onto Trevigen Flare slides (Gaithersburg, MD, USA) for each of the three damage assessments. Slides measuring in vivo damage were immediately exposed to lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris base, 1% Triton X-100, 10% DMSO, pH 10.0). Challenge slides were exposed to 200µM H2O2 for five minutes prior to lysis. Repair slides were exposed to 200µM H2O2 for five minutes followed by incubation at 37°C and 5% CO2 in RPMI complete media for one hour prior to lysis. After lysis, cells were digested by Endonuclease III and denatured prior to a 20 minute electrophoresis at 35 V and 300 mA in an 1X alkaline buffer (10 N NaOH, 200 mM EDTA, pH>13.0). During the electrophoresis, undamaged DNA remains consolidated in the form of a distinct head while degraded strands migrate out of the head leaving an elongated tail. Following electrophoresis all slides were neutralized (0.4 M Tris base, pH 7.5), washed in 100% EtOH, air dried, and stored at room temperature until visualization. To visualize, slides were stained with DAPI and imaged via confocal microscopy. DNA damage to erythrocytes was quantified using Comet Score software.