Student Handout - Bioinformatics Activity Bank
advertisement

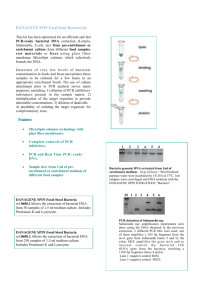
What Was In That Sandwich? By Bronston Sterner This activity is a slight modification of a protocol available at http://faculty.unlv.edu/wmojica/PCR_LAB2.htm Purpose: To use different forms of technology to isolate and manufacture more of the prokaryotic DNA found in a sample to determine a specific pathogen causing the illness of a patient. Scenario: Scott has gone to the local sub shop and purchased a turkey sub with lettuce, tomato, spinach, cheese and their special sauce on it. He had the sub back at his house along with a glass of water for lunch. Scott cleaned up his mess (he is a sloppy eater) and then went back to work outside planting the flowers his wife had bought at the store. Five days later Scott became ill and was wondering what could have caused this. He went to the doctor next day because he was becoming dehydrated due to the events which were leading to a loss of fluids he was experiencing. The doctor believes this could be some type of food poisoning but needs to identify the specific pathogen causing the illness. The doctor is relying on you to identify the specific pathogen. Time is of the essence. Background Information: The PCR (Polymerase Chain Reaction) process, was invented by Kary Mullis in 1984 and has been used by various individuals for identifying molecular sequences (Fester). Previously if only a small amount of DNA was available, tests could sometimes not be done due to the small amount available. But with new technology this issue can be addressed. Once you have your DNA sequence you simply need to make more of it so that the DNA can be detected. This process of making more is referred to as amplification. To amplify DNA you need a pair of primers (one at the beginning and one at the end), a bunch of free nucleotides (dNTPs), and a builder to assemble the free nucleotides to the DNA strand called DNA polymerase. After you have the components then a cyclical process occurs involving heat and a cooling time. The three steps are below in the diagram and include: Heating of the solution so the DNA strand can be separated. (Step 1) Then the solution will be cooled so the primers can attach to the correct location on the DNA strand. (Step 2) Finally the solution is heated up so that DNA building can take place by the DNA polymerase. (Step 3) When all three steps are completed this is referred to as a cycle. So if there were 5 DNA strands in the solution at the beginning, after one cycle there would be ________ DNA strands. After the fifth cycle occurs, there will be ________ strands of DNA. To get enough DNA to test we will be running about 25-40 cycles. IMPORTANT REMINDERS ALWAYS keep PCR reagents on ICE. Remember the centrifuge tubes are sterile so as soon as you are ready to start making the cell suspensions get your tubes and close them so that nothing contaminates your centrifuge tubes. Gloves are good to use to keep your tubes from getting contaminated. Make sure you do not cross contaminate with the tips of the micropipettes. Micropipettes come in different sizes and so do the pipette tips make sure you are using the correct pipette and tip for the amount you need to pick up. The p20 micro pipette is only accurate for picking up 2μL - 20μL A p200 micro pipette is only accurate for picking up 20μL - 200μL The p1000 micro pipette is only accurate for picking up 100μL - 1000μL Remember, if you are not sure about the last thing you did with your pipette tip then you should just get a clean one. Dirty pipette tips should be placed in waste containers and not touched. The best thing to do is to label the micro centrifuge tubes (LABEL EVERYTHING). Part I: Amplification (Day 1) 1. Get a sterile PCR tube close it. LABEL it with your group number, and then open it to pipette 5μL of your (well mixed) eukaryote n prokaryote cell suspension. Close and place PCR tube in ice. PCR Protocol: 1. Label a PCR tube with your group number and a + symbol and another PCR tube with your group number and a - symbol. Set these to the side with lids closed. 2. In your Pink tube you will be adding the below materials to it. This is called your MASTER MIX container. A. Pipette 50μL of Promega Master mix (Wards Supply Co) into the Master Mix container. B. Pipette 1.0μL of 1494 R primer into the Master Mix container C. Pipette 1.0μL of 530 F primer into the Master Mix container D. Pipette 38 μL of distilled water into the Master Mix container 3. Pipette 40μL of Master Mix into the + PCR tube and 2.5μL of your DNA also into the + PCR tube. Mix using the vortex machine for 2-3 seconds. Then place this on ice. 4. Pipette the remainder of the Master Mix into the negative PCR tube and 2.5 μL of distilled water into the negative PCR tube. Mix using the vortex machine for 2-3 seconds. Then place this on ice. 5. Each group will take the 2 PCR tubes back to the instructor when called. Work on the questions below until that time. Questions: 1. Why would samples not have evidence of the R16 gene in them. Provide 2 reasons as to why this may not work besides student error in performing the experiment. 2. Provide 3 other examples, besides bacterial identification, of ways that PCR could be used to identify a certain gene in an organism 3. Why do you think there is a range of 25-40 cycles? Why can’t the cycle number be more precise? After the PCR: As we said before to see your PCR results you need to run a gel. Whenever you need to work with the gel; gloves MUST be worn. To have a comparison to the DNA Migration we are going to be using a marker. It’s not a crayola one but is referred to as a DNA marker which will show you where the 1500 Kb is located at on the gel. The ladder is a known mixture of nuclease free H20 and dye. Results of the unknowns can then be compared to the known marker. Be careful when you load your solutions into the gels. They are like jello and can be broken very easily. You want to place the tip of the pipette above the well but below the buffer solution. You should not touch the gel with the pipette. Push the plunger down slowly to dispense the solution. It is heavier than the buffer solution so it will fall to the bottom of the well. If done correctly your colored solution will be held in the rectangular well. Okay so we have replicated our DNA from the bacteria and it appears that nothing is in our tube. But to test this we need to run a gel using the process of electrophoresis. This is similar to the chromatography activity, except we are using a gel. This will help to determine if bacterial DNA is found in our sample or not. Part II: Gel Electrophoresis - DNA separation (Day 2) 1. Add 5 μL of loading dye to the positive PCR tube and 5 μL of loading dye to the negative PCR tube. 2. Then take 15 μL of the ladder and pipette into the first well on the gel. Remember to slowly release the entire solution into the well. 3. Then take your + PCR solution and pipette 15 μL of the positive PCR tube into the second well. Remember to slowly release the entire solution into the well. 4. Then take your - PCR solution and pipette 15 μL of the negative PCR tube into the third well. Remember to slowly release the entire solution into the well. 5. The second group at your table will be using wells 5(+) and 6(-) for their solutions. 6. Once all groups have put their PCR products into the appropriate wells the lid can be placed on the container with the colored electrodes being matched up. This should be checked by the instructor before the machine is turned on to 150V. We will read the gel the next day. Part III: DNA Separation (Day 3) Gel Outcomes: * A positive result for bacteria will have the 16S Gene deposited at 1500 kB, such as the cartoon rendition in Well 2. Which Bacteria are Present? The DNA was analyzed and the nucleotide sequences were identified. These sequences follow the instructions and are to be used to identify the bacteria could have caused the illness. Remember not all bacteria are pathogenic. You will need to find out what the six bacteria are by using their nucleotide sequence and then research the scientific names to determine if they are pathogenic or not. If they are pathogenic, then you must research to see if they could cause some type of food poisoning associated with the patient Scott. Computer Analysis of Sequences 1. Type in the word BLAST into Google. 2. The first site listed has the link blast.ncbi.nlm.nih.gov/ - click on this to take you to the website you will be using. 3. Click on nucleotide blast, which will take you to the page to help identify your bacteria. You will want to copy the nucleotide sequences which were found for the bacteria and paste the entire sequence into the box labeled Enter accession number(s), gi(s), or FASTA sequence(s) 4. Go down to Choose Search Set and under database select nucleotide collection 5. Then click on 6. This may take some time before possible choices are listed. 7. Read through the list to determine which specific bacteria is the most probable one based upon Max Score and Query coverage. The closer it is to 100% the more likely it is the bacteria. You are only concerned with the genus and species, not all the other information after it. 8. After you have your list of probable bacteria, have it checked by your instructor. 9. Then research the bacteria you have identified to see which one may have caused the illness in our patient Scott based upon symptoms and what he ate. Number Scientific Bacterial Name Star the One Which Is The Pathogen 1 2 3 4 5 6 Based upon your research justify why you believe the _____________________________ bacteria is the one which caused Scott to become ill. _____________________________________________________________________________________ _____________________________________________________________________________________ _____________________________________________________________________________________ _____________________________________________________________________________________ _____________________________________________________________________________________ _____________________________________________________________________________________