DOI: 10
advertisement

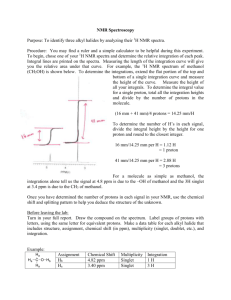
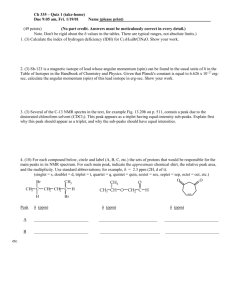
Biomimetic nanoparticles with polynucleotide and PEG mixed-monolayers enhance calcium phosphate mineralization Kayla B. Vasconcellos, Sean M. McHugh, Katherine J. Dapsis, Alexander R. Petty, and Aren E. Gerdon* Emmanuel College, 400 The Fenway, Boston, MA 02115 (USA) E-mail: (gerdoar@emmanuel.edu) Phone: 617-735-9769 Fax: 617-735-9877 Supporting Information S1. TEM and histogram of T-MPC. S2. Proton NMR and UV absorbance characterization of tiopronin and mixed monolayer nanoparticles. S3. DLS histograms of T-MPC calcium phosphate mineralization. S4. Complementary molybdate analysis data. 1 S1. TEM and histogram of T-MPC. Electron microscopy shows spherical particles with a broad size distribution and average diameter for the gold core of 4+2 nm. 2 S2. Proton NMR and UV absorbance characterization of tiopronin and mixed monolayer nanoparticles. Multiple proton NMR spectra are displayed below. The tiopronin small molecule shows sharp peaks relating to CH 3 (1.5 ppm, doublet), CH2 (4.0 ppm, singlet), and CH (3.7 ppm, quadruplet) protons. Corresponding, but broad peaks in the T-MPC indicate the formation of the nanoparticle and are due to the combination of many tiopronin molecules per nanoparticle, each in a slightly different local environment. The lack of sharp peaks indicates a lack of unattached tiopronin and demonstrates the purity of the sample. Briefly, the calculation of 250 tiopronin molecules per nanoparticles comes from a combination of TEM data (S1, above), and previous thermogravimetric analysis studies (Gerdon, et al. 2005). This diameter MPC (~4 nm) is typically 80% gold by mass and 20% organic ligand by mass, according to thermogravimetric analysis (TGA). Based on the average diameter of 4 nm and the face-centered cubic packing of gold, the number of gold atoms per MPC can be calculated. Using this number of gold atoms, the mass of the core is calculated and based on thermogravimetric analysis percent composition, the mass of tiopronin is calculated. This mass provides the approximate number of tiopronin ligands per gold core. Proton NMR spectra, below, were taken to confirm the presence of the place-exchanged ligand on the surface of the T-MPC. NMR relative integral values were used to determine the monolayer composition of each PEGMPC. In the broad MPC proton NMR spectra, one peak is observed for the CH3 protons (1.7 ppm) and one peak is observed that includes the overlapping CH2 and CH protons (4.0 ppm). The sum of the integration value at 1.7 ppm should equal the sum of the integration values at 4.0 ppm in T-MPC since each peak is indicative of 3 protons. The PEG molecule has 44 CH2 protons which result in an NMR shift at 3.7 ppm in both the free PEG molecule and in PEG that has been exchanged onto T-MPC. The PEG protons therefore overlap with the protons from tiopronin near 4.0 ppm. After place exchange, the integration value for all peaks ranging from 3.54.5 ppm should increase relative to the integration peak for 1.7 ppm. Also, place exchange reactions occur in a one-to-one stoichiometry, so that when one PEG molecule attaches to the MPC, one tiopronin molecule dissociates. The following equations represent this and were used to calculate the number of PEG molecules per cluster: [250 tiopronin per MPC] × [3 CH3 protons per tiopronin] = [750 total protons in CH3 peak at ~1.7 ppm] (1) [250 tiopronin per MPC] × [3 CH2 and CH protons per tiopronin] = [750 total protons in CH2 and CH peaks from ~3.5-4.5 ppm] (2) Integration Value (3.5−4.5 ppm) Integration Value (1.7 ppm) = 750−3x+44x 750−3x (3) where, the x indicates the number of PEG molecules exchanged onto the surface of the MPC. Because characteristic peaks for tiopronin CH2 protons and PEG CH2 protons appear in the same region, their normalized peak area accounts for both sets of protons. In the representative proton NMR below, the integral 3 value from 3.5-4.5 is equal to 1.91 compared to 1.00 at 1.7 ppm, indicating the presence of 15 PEG molecules and 235 tiopronin molecules. Since the NMR spectra of DNA is significantly more complicated than that of PEG, UV absorbance rather than NMR was used to quantitate the DNA after place exchange. DNA has a distinctive peak at 260 nm, as shown below. After purification by dialysis, all DNA present is attached to the MPC. Therefore, the absorbance at 260 nm was used to determine the concentration of DNA, based on a known molar absorptivity. 4 S3. DLS histograms of T-MPC calcium phosphate mineralization. Early time-point histograms show large polydispersity and multiple size range particles likely due to mineralization at various stages of nucleation and growth. After 10 min, polydispersity has decreased as nucleation slows and particles grow at similar rates. By 60 min, smaller particles have grown and aggregated to very large sizes. 5 S4. Molybdate blue assay. Absorbance spectroscopy was used to quantify the total phosphate content in mineralized samples (Figure below, left). Briefly, MPC samples (50 L, 0.17 mg/mL, filtered) were combined with 500 L each of 10.0 mM calcium chloride and 4.0 mM sodium phosphate pH 7.4 and vortexed to mix. Samples were allowed to react undisturbed for 60 minutes and then centrifuged at 12000 rpm for 60 seconds to collect and wash the precipitate with DI water. Each sample was washed with DI water and centrifuged a total of five times. Mineralized material was then dissolved in 1000 L of 0.05 M HCl, treated with 250 L of 2 mM EDTA, combined with 250 L of 4 mM ascorbic acid, and combined with 500 L of 5.0 mM ammonium molybdate. After reaction for 15 min, absorbance measurements were taken with an Agilent 8453 UV-Visible Spectrophotometer at 650 nm and compared to a standard curve. A complementary experiment (Figure above, right) was completed to analyze the effects of washing and centrifuging on the error associated with the method of analysis used above. This experiment was set up exactly as above, but instead of collecting and analyzing the mineralized pellet, the supernatant was collected and analyzed via molybdate assay instead. This removed the necessity of washing the pellet and required only one centrifugation step. This method provided the mole amount of phosphate remaining after mineralization as well as the mole amount of phosphate removed from solution via precipitation. Unfortunately, the percentage of phosphate removed from solution was small and the error associated with the experiment was still relatively large at ~12% error. This points to variability in the molybdate assay rather than to variability in the mineralization process itself. The data correlates very well with the previous data, in term of magnitude of phosphate consumed. 6