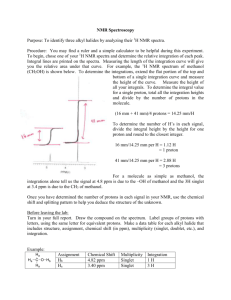
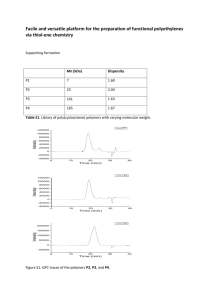
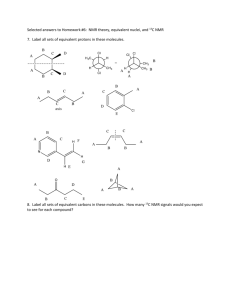
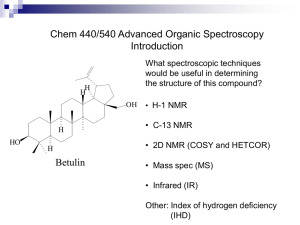
NMR - University of Puget Sound
advertisement
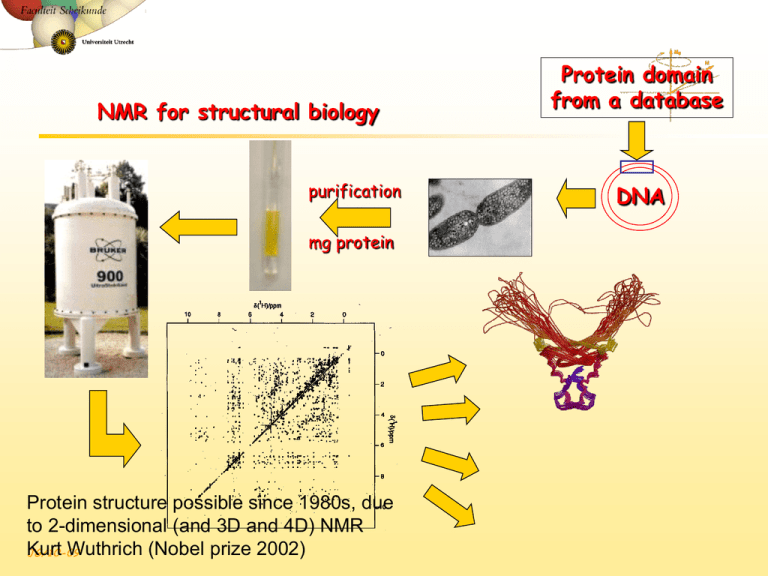
NMR for structural biology purification mg protein Protein structure possible since 1980s, due to 2-dimensional (and 3D and 4D) NMR Kurt Wuthrich (Nobel prize 2002) JG/10-09 Protein domain from a database DNA NMR structure determination steps • NMR experiment • Resonance assignment (connect the spin systems with short-range NOEs) • Structural restraints • • • Structure calculations • • JG/10-09 Distances (from NOEs) torsion angles (from J coupling) Conformation of polypeptide that satisfies all distance restraints Structure validation (cross-check your data) Step 1: The resonance assignment puzzle 750 MHz 1H NMR spectrum of the SH3 domain of the tyrosine kinase FYN Hydrogen atoms JG/10-09 Ribbon representation Solutions to the Challenges • Increase dimensionality of spectra to better resolve signals: 1234 • Detect signals from heteronuclei (13C,15N) • • JG/10-09 Better resolved signals, different overlaps More information to identify signals HNCO JG/10-09 Step 1: 2D COSY spectrum. Protons transfer magnetization to 3-bond coupled protons (J-coupling) Crosspeaks at chemical shift of correlated protons. Assign amino acid spin systems (3-bond coupled protons)! JG/10-09 2D NOESY spectrum: through-space <5Å distances (H-H) JG/10-09 Step 2: Structure calculation using distance matrix • • Nuclear Overhauser Effect (NOE) crosspeaks for all protons <5Å apart (through space!) With assignments, use NOEs to calculate structure of protein (distance matrix) Tertiary Structure Sequential Intraresidue JG/10-09 A B C D Medium-range (helices) •••• Z NMR Structure ensemble • 20 structures all satisfy observed NOE (distance) data • Some regions of protein are more dynamic, and the NMR structures show the range of conformations that the protein samples. JG/10-09 A folded protein is easily recognized, and H/D exchange tells about hydrogen bond stability JG/10-09 Protein domain Globular protein tertiary structure Sidechain location vs. polarity -Nonpolar residues in interior of protein (hydrophobic effect promotes this, as well as efficient packing of those sidechains) -Charged polar residues on protein surface (immersing charge in anhydrous interior is energetically unfavorable) -Uncharged polar groups occur in both places (hydrogen bonding and electrostatic interactions inside the protein “neutralize” their polarity) Amphipathic helices, sheets Protein interiors compact (more efficient packing than organic molecule crystals!) -however, they have low-energy arrangements of sidechains (no steric clashes) -exclude water (where present it often makes specific H-bond “bridge”) -maximize vdW surface complementarity These low-energy characteristics have evolved…(If a more stable protein helps it function and the organism survive, then the amino acids conferring the most stability will be selected for.) a-helix b-sheet purple nonpolar Globular protein structure4 Sample problems: 1) Estimate the number of amino acids in this protein. 2) What are reasonable amino acid sidechains for the inner and outer faces of these helices? (Just do the first three turns of the red one) Draw or name 3 interior and 3 exterior a.a. for each helix. 3) What a.a. sidechain(s) can coordinate the heme iron atom? exterior face interior face Double-resonance experiments increase resolution/information content Correlates proton chemical shifts with chemical shifts of other NMR nuclei such as 15N (needs labeling!) or 13C (1.1% natural abundance, possible with 100mM fumarate, but not protein…aggregation!) R R -15N-Ca-CO-15N-Ca H JG/10-09 H 15N-1H HSQC