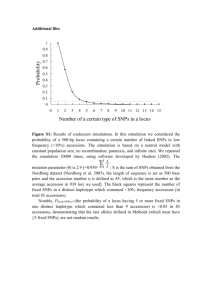
README

Notes on running our simulation model:
Run the script in the file named “kardos_HFC_ID_simModel_11September2013.r”. Carefully follow the directions given at the beginning of the script.
As the simulation script is written here, there is a single output space delimited text file for each simulation repetition:
The file contains all of the pedigree information, pedigree inbreeding coefficients, the proportion of the genome that is IBD (F), and genotypes at simulated SNPs and microsatellite loci. The number of columns will depend on the input parameters for the simulation run. However columns 1-6 contain the generation, ID, sex (1=male, 0=female), parent number 1 ID, parent number 2 ID, immigrant status (1= immigrant, 0 = resident), and F, respectively. The file will then contain information on the pedigree inbreeding coefficient – one column for each designated depth of pedigree the user specified in the simulation run. The file will then contain columns for multiple locus heterozygosity calculated at the simulated SNP loci – one column for each number of SNPs specified by the user for the simulation run.
The there will be one column for each of the SNP loci indicating whether the individual is heterozygous
(1) or homozygous (0) the locus. Next, there will be a single column for each simulated microsatellite locus, each indicating if the individual is heterozygous or homozygous at the locus (as with SNPs). Lastly, there will be columns indicating multiple-locus heterozygosity at the simulated microsatellite loci.
The data in this file can be used to test for inbreeding depression using F or individual heterozygosity.