2012a Jan Answer Guide
advertisement

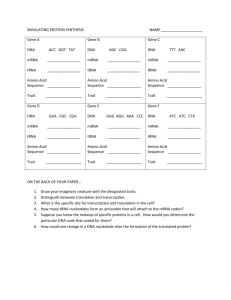
School of Science Hamilton Campus Session 2011 – 12 Trimester 1 Module Code: BIOL09020 PURE AND APPLIED GENETICS ANSWER SCHEME Date: January 2012 Time: Duration: Answer THREE questions, at least ONE from each section Page 1 of 10 Continued overleaf BIOL09020 1. a) Answer Scheme January 2012 Discuss the features of the plasmid pUC18/19 (Figure 1) which make it a good vector for cloning DNA in bacterial cells. (6 marks) Answer Key features: As a plasmid it can be replicated inside the host cell. Often high copy number. Contains multiple unique cloning sites (single restriction sites to insert fragment). Selectable markers to enable transformed cells to be identified. May also include: Relatively small and easily isolated from the cell. Cells can be forced to take up the plasmid in vitro. b) Describe the steps you would perform to clone a DNA fragment in E.coli using the vector in figure 1. Answer The DNA to be cloned and the plasmid are cut with the same restriction enzyme. The enzyme should cut uniquely within the MCS and at either end of the DNA fragment. Enzymes that produce sticky ends are preferred. The plasmid could be treated with alkaline phosphatase prior to ligation to reduce the chance of vector religation. Cut plasmid and fragment are mixed in a ligation reaction with DNA ligase. The complementary sticky ends should base pair and ligase forms the phosphodiester bonds. The ligation mix would be used to transform bacterial cells, E.coli. Probable method would involve calcium chloride and heat shock (explanation of principles of technique). Control with no DNA included. Both the ligation reaction and the transformation reaction are inefficient resulting untransformed cells, cells containing plasmid only with no inserted DNA and cells containing the desired recombinant plasmid. The desired cells are selected – after transformation cells are plated onto agar containing ampicillin and X-gal. Ampicillin will kill all untransformed cells which are not resistant to the antibiotic while all cells containing the plasmid will survive. Those containing plasmid only or recombinant plasmid are selected using X-gal. The lac Z gene in the plasmid codes for galactosidase which converts the substrate X-gal into a blue product making those colonies containing plasmid only blue. In the recombinant plasmid the foreign DNA fragment is inserted into the lac Z gene inactivating it therefore it does not produce B gal cannot break down the substrate and therefore the colonies remain white/cream. To confirm the insert is present a plasmid miniprep is used to isolate the plasmid. Cut the isolated plasmid with restriction enzymes appropriate to show the presence of the fragment and its orientation. Run on agarose gel and visualise. Page 2 of 10 (14 marks) BIOL09020 Answer Scheme January 2012 Figure 1 Page 3 of 10 Continued Overleaf BIOL09020 Answer Scheme January 2012 2. You are studying a gene coding for an enzyme that breaks down toxins which are harmful to the cell. a) Describe how you would use the polymerase chain reaction (PCR) to amplify the gene from genomic DNA. (12 marks) Answer Answer should include key points as follows: Basic components are DNA template (Generally double stranded DNA, no need to be purified, continuous sequence spanning the region to be amplified), DNA Polymerase (thermostable), Oligonucleotide primers (Two synthetic oligonucleotides (20–30bp) that hybridise to opposite strands of the DNA and flank the DNA of interest allowing extension to the 3’ end), Deoxynucleotide triphosphates, Reaction buffer, Magnesium. PCR is a repeated cycle of 3 steps each cycle the number of specific DNA templates doubles. Denaturation (94oC) – Heat is used to break the H bonds between strands denaturing the DNA allowing it to be used as a template. Annealing (40 – 60oC) – the primers anneal to the template, exact temperature depends on the primer. Extension (72oC) – DNA Polymerase adds nucleotides to the 3’end of the primer. Repeated at least 30 times. b) Outline how the basic PCR technique can be modified to determine if the gene is expressed after exposure to toxins. (8 marks) Answer Reverse transcriptase PCR should be described. RNA is isolated from the cell and reverse transcribed to cDNA using the enzyme reverse transcriptase. Oligo dT can be used as the primer which would produce cDNA from mRNA. The presence of cDNA specific for the enzyme can be determined by using cDNA as the template in PCR with primers specific for the gene of interest. A product will only be produced if the mRNA was present originally. May suggest the amount of expression could be determined using qPCR. Page 4 of 10 Continued Overleaf BIOL09020 3. Answer Scheme January 2012 Two types of genes, oncogenes and tumour suppressor genes have been shown to play a role in the development of cancer. Discuss the normal function(s) of these genes in the cell and examples of mutations that can lead to cancer. (20 marks) Answer Answer might include the following points. Oncogenes are cancer causing genes ie. expression results in cancer. These are mutated versions of the normal gene (protooncogene) genetically dominant; one copy can change the cells behaviour. They are cellular genes whose function is to promote normal growth and division of cells. Expression of these genes is tightly controlled in a normal cell. Changes to the genes may increase their expression resulting in increased cell division. Oncogenes may be cell surface receptors, growth factors, molecules involved in transmembrane signalling, second messengers or nuclear proteins. Both qualitative and quantitative effects in protein expression should be discussed with examples. Burkitts lymphoma is an example of a quantitative change translocation alters transcription of the myc oncogene on chromosome 8 to chromosome 14, 22 and 2 in regions where there are immunoglobulin loci. These genes are actively transcribed. There are two possibilities a repressor region is destroyed or myc is influenced by the immunoglobulin enhancer both result in increased expression. C-myc is a DNA binding protein. Qualitative change example could be EGF receptor and erbB. ErbB is a truncated EGF receptor which is permanently switched on. There are many other examples that could be used. Tumour suppressor genes are genes whose normal function is to inhibit the cell cycle and cell division. The gene coding for p53 is mutated in the majority of cancers implying that the loss of the normal gene product is associated with malignancy. p53 plays a central role in controlling the cell cycle and loss of function leads to uncontrolled cell growth. It is a transcription factor, which activates the expression of cellular genes whose products cause the cell cycle to arrest in G1. Loss of p53 means these genes are not activated and the inhibition in G1 is removed. Page 5 of 10 Continued Overleaf BIOL09020 Answer Scheme January 2012 4. a) Compare and contrast the content and organisation of the genome in prokaryotes and eukaryotes. (10 marks) Answer A typical 50kb segment of human genomic DNA may contain: One gene coding for a protein. Genes coding for proteins contain introns and exons (on average 9 per gene). During transcription the entire gene is copied to pre mRNA but introns are subsequently removed by splicing to produce mRNA which is used to direct protein synthesis. Gene segments Pseudogene – non-functional copy of a gene. May have been a gene inactivated by mutation which has accumulated further mutations or derived from mRNA. A DNA copy of the mRNA is made by reverse transcriptase. The copy is then inserted into the genome but is inactive as it lacks introns and upstream sequences. Interspersed repeat sequences that occur in several places throughout the genome. Microsatellites – sequences that are repeated several times. 50% non coding, non-repetitive single copy DNA with no known function Bacterial DNA is much smaller and generally circular. It is much more compact, with far more genes in a smaller space and very few repetitive sequences. Very little non-coding DNA ~ 11%. No introns. Genes are often very close, arranged in operons where several genes are transcribed as a single mRNA from one promoter. Sense sequences are on both strands. Bacteria often contain plasmids. Plasmids are small extra-chromosomal DNA that contain genes. The genes give bacteria additional properties e.g. antibiotic resistance but are dispensable. b) Outline why the ends of linear chromosomes get shorter during successive rounds of replication in eukaryotes. (4 marks) Answer The chromosome becomes shorter once the last primer is removed from the end. DNA polymerase has no 3’ end to add on to complete the gap. Diagrams might help to explain this. Page 6 of 10 Continued Overleaf BIOL09020 c) Answer Scheme January 2012 Discuss the effect telomere shortening has on the cell and describe the mechanism some cells have developed to overcome. (6 marks) Answer The chromosome shortens every time the cell divides. Shortening limits the number of cell divisions as eventually genes become damaged and cells can no longer divide. The presence of active telomerase prevents shortening. Telomerase provides a short molecule of RNA to allow extension of the 3’end. There is then sufficient length to prime the 5’ end once the primer is removed the 5’end is the correct size. (important in stem cells and cancer cells). Page 7 of 10 Continued Overleaf BIOL09020 5. Answer Scheme January 2012 Discuss the events that occur at the level of transcription when E.coli is grown on a mixture of glucose and lactose. (20 marks) Answer E. coli will use glucose as a carbon source for growth first. Glucose levels will decline while lactose remains constant. During the decline the bacteria show exponential growth. Growth is followed by a lag after which lactose levels decline with a second exponential growth suggesting lactose is used as a carbon source. When the lactose is depleted the bacteria enter a stationary phase. In the absence of lactose the lactose repressor is bound to the operator preventing binding of RNA Polymerase and transcription of the operon. The repressor occasionally detaches allowing a few molecules to be transcribed. When the bacteria encounter lactose the low level of enzymes allow it to be transported into the cell and metabolised to glucose and galactose. An intermediate in this reaction is allolactose, which binds to the repressor, changes its conformation so it can no longer bind to the operator. RNA Polymerase can then bind to the promoter and transcribe the 3 genes. Transcription is still low without the activation of CAP. Catabolite activator Protein This protein binds to sites upstream of operons including the lactose operon and increases the efficiency of transcription initiation. When E.coli is grown in the presence of glucose as well as lactose the glucose acts to override the lac operon. Glucose controls the activity of adenylate cyclase an enzyme that converts ATP to cAMP. Glucose inhibits the enzyme decreasing cAMP levels. The CAP can bind to DNA only in the presence of cAMP therefore in the presence of glucose binding is inhibited and the lac operon is only transcribed at a very low level. Diagrams showing the arrangement of the operon may be useful. Page 8 of 10 Continued Overleaf BIOL09020 6. Answer Scheme January 2012 Three types of RNA are involved in Translation (Protein synthesis) discuss the role and interaction of all three. (20 marks) Answer This is an example of what might be included but alternatives are acceptable. The function and role of each RNA molecule should be described in detail. mRNA carries the code to be translated copied from DNA. It is read 5’ to 3’ in groups of 3 nucleotides, codon. rRNA and proteins make up the ribosome, site of protein synthesis. Ribosomes consist of two subunits, small and large which are separate in the cytoplasm when not involved in protein synthesis. The small subunit and IF3 attach to mRNA. Attachment is to the ribosome-binding site (Shine Dalgarno sequence). This is a sequence on the mRNA upstream of the start codon which is complementary to a sequence on the 3’ end of 16s rRNA. Attachment positions the small subunit over the initiation codon, AUG. The structure of tRNA should be drawn or described. Two key areas highlighted – anticodon – triplet of nucleotides that base pair with the codon on mRNA, specific interaction involving base pairing. A diagram could be used to show pairing and direction of RNA’s. Wobble could be described. The codon- anticodon interaction brings the correct tRNA. - Amino acid acceptor stem, 3’ end has CCA where amino acid is attached. Addition of amino acid is catalysed by aminoacyl tRNA synthetases. Reaction can be described, isoaccepting tRNAs. The enzymes have high fidelity for their tRNA structurally similar amino acids can be edited. Some amino acids are attached and modified. The key is to have the correct amino acid attached. When the large subunit attaches to form the complete ribosome there is space for 2 tRNA molecules plus exit site in the cavity between the subunits. The small subunit has the codonanticodon interaction and the large subunit has the aminoacyl end. The initiator tRNA occupies site P, peptidyl site. The second site A, aminoacyl has the second codon exposed and the tRNA complementary to the codon is brought to the site by elongation factors, which ensures the correct amino acid is attached. Contacts between tRNA, mRNA and 16sRNA ensure correct tRNA is accepted, correct base pairs must be formed between all 3 nucleotides. A peptide bond is now formed between the amino acids held by the 2 tRNA’s. Peptidyl transferase catalyses this reaction and the release of the amino acid from the tRNA in site P. In bacteria this activity is part of the 23s rRNA and is therefore a ribozyme. The reaction requires energy. Page 9 of 10 Continued Overleaf BIOL09020 Answer Scheme January 2012 All the following occur at once: Ribosome moves along 3 nucleotides to expose third codon in site A. The tRNA carrying the dipeptide moves from site A to P. The empty tRNA in site P moves to a third exit site in bacteria or is ejected in eukaryotes Energy is required. Electron microscopy suggests that the 2 ribosomal subunits rotate slightly in opposite directions creating a space enabling the ribosome to slide along the mRNA. The empty A site is filled with the appropriate tRNA and the elongation cycle continues to the end of the open reading frame. Diagrams would be good. END OF QUESTION PAPER Page 10 of 10