DNAJ paper (NBA)
advertisement

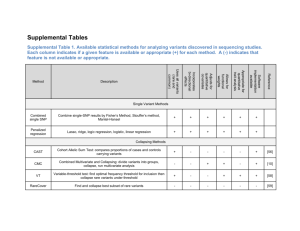
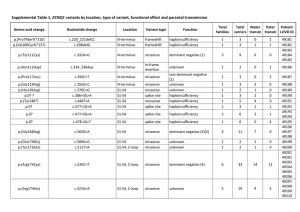
DNAJ mutations are rare in Chinese Parkinson’s disease patients and controls Jia Nee Fooa, Herty Lianya, Louis C. Tanb, Wing-Lok Aub, Kumar-M. Prakashb, Jianjun Liua, Eng-King Tanb,c * a. Human Genetics, Genome Institute of Singapore, A*STAR, Singapore 138672. b. Departments of Neurology, Singapore General Hospital, National Neuroscience Institute, Singapore 169108 c. Duke-NUS Graduate Medical School, Singapore Corresponding author: Eng-King Tan, Department of Neurology, National Neuroscience Institute, Duke-National University of Singapore Graduate Medical School, Singapore General Hospital, Singapore 169108; tan.eng.king@sgh.com.sg ; +65-63214006 1 Abstract Mutations in DNAJC13, DNAJC6 and DNAJC5 have been implicated in Parkinson’s disease (PD). To determine if rare coding variants in these genes play a role in PD risk in the Chinese population, we sequenced all coding exons of the three genes in 99 early-onset PD cases and 99 controls, and genotyped 8 missense variants in another 711 PD cases and 539 controls. Besides two common missense variants that did not show association with PD, the remaining missense variants were extremely rare (<0.5%), found in healthy population controls and did not show enrichment in PD cases. Our results suggest that missense mutations in DNAJC13, DNAJC5 and DNAJC6 do not play a major role in PD in the Chinese population. 1. Introduction DNAJC13 has recently been identified as a novel gene implicated in late-onset Lewy body Parkinson’s disease (PD). The rare missense mutation p.Asn855Ser was identified through linkage analysis and exome sequencing of a large Saskatchewan Mennonite family, in which twelve out of 57 members had previously been diagnosed with PD (Trinh & Farrer 2013). Other genes in the DNAJ family have previously been implicated in familial neurodegenerative disorders, including DNAJC6 in recessive juvenile Parkinsonism (Edvardson et. al. 2012) and DNAJC5 in dominant adult-onset neuronal ceroid lipofuscinosis (Kufs disease) (Cadieux-Dion et. al. 2012). 2. Methods To study if rare coding variants in these genes play a role in PD risk in the Chinese population, we sequenced all coding exons of the three genes in 99 early-onset (<55 years; age 54.7 ± 7.1 years, onset 48.3 ± 5.8 years, 67% male) and familial (23%) PD cases and 99 elderly, healthy controls (age 71.9 ± 10.8 years, 61% male) from Singapore. We further analyzed 2 recurrent missense variants that were genotyped as part of the Illumina HumanExome BeadChip Exome_Asian array in an additional 711 cases with typical late-onset PD (>55 years; age 69.4 ± 9.2 years, onset 65.3 ± 9.2 years, 51% male) and 539 healthy controls (age 51.7 ± 11.6 years, 57% males) from Singapore. Patients were diagnosed with PD using the UK Brain Bank Criteria and population controls were recruited. Cases and controls were confirmed to be well-matched in a principal components analysis using genome-wide SNPs from the Illumina array. All subjects gave informed consent and the study received approval from the institutional ethics committee. Allele frequencies in cases and controls were compared using Fisher’s exact tests. We used the SIFT algorithm to predict the effects of missense variants (Ng & Henikoff 2003). 3. Results & Discussion None of the reported disease causing mutations was identified in any of the three genes in the sequencing analysis of 198 samples. In DNAJC13 (NM_015268.3), we identified three missense mutations and six silent mutations. Of the three missense mutations, one was common (p.Ala1463Ser; >20% frequency in 1000 genomes populations and 99 controls), one was rare (p.Gly368Cys; predicted damaging) and found in one control subject, and one was rare (p.Met2225Ile; predicted tolerated) and found in only one case subject (Supplementary Table 1). p.Met2225Ile is also present in 1000 genomes populations, with frequencies ranging from 0.7% (Asians) to 6% (Americans). In DNAJC6 (NM_014787.2), we identified three missense variants and seven silent mutations (Supplementary Table 1). Two of the missense variants were rare (<5% in 99 controls and 1000 genomes populations), with one found in two control subjects (p.Asn469Ser; predicted tolerated) and one found in a single case subject (p.Gly597Asp; predicted damaging). No nonsense, splice-site or frameshift mutations in DNAJC6 were identified in our samples, and 3 none of the PD cases were homozygotes or compound heterozygotes for missense variants. In DNAJC5 (NM_025219.2), no missense variants and only two silent mutations were identified (Supplementary Table 1). None were located near splice sites. We further genotyped p.Met2225Ile, p.Ala1463Ser and five other missense mutations (present on the Illumina exome chip) in DNAJC13, as well as p.Ser671Asn and p.Asn469Ser in DNAJC6 in 711 late-onset/sporadic cases and 539 healthy controls. Neither of the two common variants (p.Ala1463Ser, p.Ser671Asn) showed association with PD, even though we had >75% power to detect a variant with a frequency of 10% and odds ratio of 1.4 at the significance threshold of alpha=0.05. All the other variants were extremely rare (<0.5%), found in healthy controls and none showed evidence of enrichment in PD cases (Supplementary Table 2). Our results suggest that missense mutations in DNAJC13, DNAJC5 and DNAJC6 are not major causes of early- or late-onset PD in the Chinese population. 4 Disclosure Statement The authors have no conflicts of interest. All subjects gave informed consent and the study received approval from the institutional ethics committee. Acknowledgement We thank National Medical Research Council, Duke Graduate Medical School and Singapore Millennium Foundation for their support. References Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013 Aug;9(8):445-54. Epub 2013 Jul 16. Edvardson, S. et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrinuncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE 7, e36458 (2012). Cadieux-Dion M, Andermann E, Lachance-Touchette P, Ansorge O, Meloche C, Barnabé A, Kuzniecky RI, Andermann F, Faught E, Leonberg S, Damiano JA, Berkovic SF, Rouleau GA, Cossette P. Recurrent mutations in DNAJC5 cause autosomal dominant Kufs disease. Clin Genet. 2013 Jun;83(6):571-5. Epub 2012 Nov 7. Ng, PC and Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2013 31(13): 3812-4. 5 Supplementary Table 1. Coding variants identified in 99 early onset/familial PD cases and 99 controls. Missense variants are shown in bold font. Position Amino Acid Substitution SIFT prediction Allele Freq Control Allele Freq Case No. of controls No. of cases chr3 132175347 p.Gly368Cys Damaging 0.00505 0 1 0 chr3 132218623 p.Ala1463Ser Tolerated 0.904 0.879 - - chr3 chr3 132257069 132153442 p.Met2225Ile p.Thr16Thr Tolerated - 0 0 0.00505 0.00505 0 0 1 1 chr3 132166266 p.Thr82Thr - 0.0960 0.121 - - chr3 132166302 p.Leu94Leu - 0 0 0 0 chr3 132172463 p.Leu255Leu - 0 0.00505 0 1 chr3 132175602 p.Leu425Leu - 0.00505 0 1 0 chr3 132230036 p.Pro1747Pro - 0.00505 0 1 0 chr1 65858222 p.Asn469Ser Tolerated 0.0101 0 2 0 chr1 65860638 p.Gly597Asp Damaging 0 0.00505 0 1 chr1 chr1 65867519 65852546 p.Ser671Asn p.Val292Val Tolerated - 0.631 0.00505 0.535 0 1 0 chr1 65855095 p.Ser393Ser - 0.00505 0.0101 1 2 chr1 65858145 p.Glu443Glu - 0.167 0.217 - - chr1 65858151 p.His445His - 0.616 0.505 - - chr1 65860660 p.Ser604Ser - 0.611 0.500 - - chr1 65860687 p.Pro613Pro - 0.803 0.753 - - chr1 65871623 p.Ser709Ser - 0.00505 0 1 0 chr20 62559773 p.Asn25Asn - 0.00505 0.00505 1 1 chr20 62560701 p.Pro48Pro - 0.0303 0.0303 6 6 chr DNAJC13 DNAJC6 DNAJC5 Supplementary Table 2. Missense variants genotyped in 711 late onset/sporadic PD cases and 539 controls position Amino Acid Substitution SIFT prediction Allele Freq Control Allele Freq Case No. of controls No. of cases Pvalue chr3 132196651 p.Ala822Thr Tolerated 0.00278 0.00211 3 3 1.000 chr3 132196920 p.Gly882Val Damaging 0.00278 0.00211 3 3 1.000 chr3 132215496 p.Arg1382His Damaging 0.000928 0.00141 1 2 1.000 chr3 132226100 p.Tyr1673Cys Tolerated 0.000928 0 1 0 0.431 chr3 132257069 p.Met2225Ile Tolerated 2 1.000 132218623 p.Ala1463Ser Tolerated 0.00141 0.892 1 chr3 0.000928 0.890 - - 0.642 chr1 65858222 p.Asn469Ser Tolerated 0.00278 0.00356 3 5 1.000 chr1 65867519 p.Ser671Asn Tolerated 0.5843 0.568 - - 0.114 chr DNAJC13 DNAJC6 6