Supplementary information Novel recurrent mutations in
advertisement

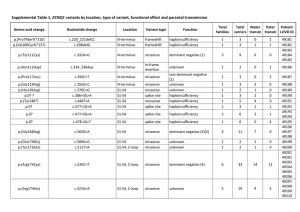
Supplementary information Novel recurrent mutations in ethanolamine kinase 1 (ETNK1) gene in systemic mastocytosis with eosinophilia and chronic myelomonocytic leukemia Lasho TL et al. Pages 2-3: Supplementary Table 1. List of somatic coding region single nucleotide variants identified in index case of aggressive systemic mastocytosis with eosinophilia Pages 4-5: Methods for whole exome-sequencing of index case for mutation identification and targeted resequencing of additional cases for mutation confirmation Page 6: Supplementary Figure 1. Nucleotide sequence conservation between species for ETNK1 gene in vicinity of region encoding for amino acid residues N244 and G245 1 Supplementary Table 1. List of somatic coding region single nucleotide variants identified in index case of aggressive systemic mastocytosis with eosinophilia Annotated Gene Mutation Type Amino Acid Change Chromosom e Position Allele Change ACAD10 AMMECR1 ATXN2L AVPR1B BMPR2 BRPF1 C4orf26 CDK19 CEACAM1 CERCAM CLIP1 COL27A1 EP400 ETNK1 EZH2 F7 FAT4 GRAMD1A GRM6 KBTBD4 KIT LCT LPHN3 MMRN2 NCAPD2 NEK8 NR5A2 PGM2 PLIN4 PLIN4 RP1L1 SCN4A SELPLG SLC17A4 SLC26A9 SLC6A2 SLFN11 STAT5A STK36 SYNE1 Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense Missense D404A S118L H871P T12P T606P L975F H97P V16G T298P Y232S E966K K452N T2216P N244S H694R V285G D2022A C456G Y487D V78G D816V T687P H1340P V433G Y1325S A306T R187G V341G C819S C819S E1300A E872G P161R I147F P746R T63P T707P V707E D1073A F8509L chr12 chrX chr16 chr1 chr2 chr3 chr4 chr6 chr19 chr9 chr12 chr9 chr12 chr12 chr7 chr13 chr4 chr19 chr5 chr11 chr4 chr2 chr4 chr10 chr12 chr17 chr1 chr4 chr19 chr19 chr8 chr17 chr12 chr6 chr1 chr16 chr17 chr17 chr2 chr6 112165822 109560947 28846647 206224474 203420204 9786696 76489546 111136293 43025485 131186822 122812709 116931191 132529360 22811995 148506431 113772775 126336183 35510328 178413880 47599367 55599321 136570175 62936235 88703243 6640096 27064863 200017395 37848566 4511475 4511474 10467709 62029022 109017650 25770436 205887987 55705945 33679962 40461400 219563485 152457885 A>C G>A A>C A>C A>C A>C A>C A>C T>G A>C C>T A>C A>C A>G T>C T>G A>C T>G A>C A>C A>T T>G A>C A>C A>C G>A C>G T>G A>T C>G T>G T>C G>C A>T G>C A>C T>G T>A A>C G>T Tlymphocyte allele fraction (control)¶ Eosinophil allele fraction (tumor) 1.90% 0.00% 2.50% 0.60% 3.00% 2.50% 0.00% 1.20% 2.50% 3.10% 3.00% 2.20% 2.90% 1.40% 3.10% 2.70% 1.40% 1.80% 1.50% 1.80% 0.00% 2.70% 2.90% 1.60% 2.90% 0.80% 0.80% 3.40% 0.90% 0.90% 1.70% 3.70% 1.30% 2.00% 0.00% 2.80% 2.20% 0.00% 0.90% 1.40% 6.20% 42.60% 5.30% 3.00% 1.60% 1.40% 2.60% 2.20% 9.20% 7.90% 2.00% 3.40% 7.40% 50.00% 97.40% 6.70% 2.70% 4.40% 2.70% 6.20% 47.10% 7.60% 1.60% 5.20% 5.40% 7.00% 1.70% 6.30% 2.70% 2.10% 0.70% 12.10% 4.90% 3.70% 35.90% 1.10% 4.10% 67.00% 2.20% 52.60% 2 TAS2R30 Missense I289T chr12 TECPR2 Missense N138S chr14 TMEM170A Missense S12A chr16 TRIM24 Missense N554T chr7 UMOD Missense L630F chr16 WDR6 Missense V884G chr3 XKR8 Missense T187P chr1 XPC Missense A425G chr3 ZEB2 Missense H95P chr2 ¶Only cases with T-lymphocyte allele fraction <4% displayed 11285978 102874889 75498465 138252356 20344671 49051528 28293082 14200109 145187383 A>G A>G A>C A>C G>A T>G A>C G>C T>G 3.00% 0.00% 3.70% 1.80% 2.70% 3.40% 0.30% 3.00% 3.40% 5.10% 53.80% 9.90% 7.20% 46.20% 5.90% 2.80% 1.60% 1.60% 3 Supplementary Methods Illumina HiSeq Sure Select XT Custom Capture and Paired End Sequencing Paired-end indexed libraries were prepared using the Agilent Bravo liquid handler following the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA). Briefly, 3 µg of target DNA was fragmented using the Covaris E210 Sonicator. The settings of duty cycle 10%, intensity 5, cycles 200, time 360 seconds generated double-stranded DNA fragments with blunt or sticky ends with a fragment size mode of between 150-200bp. The ends were repaired and phosphorylated using Klenow, T4 polymerase, and T4 polynucleotide kinase, after which an “A” base was added to the 3’ ends of doublestranded DNA using Klenow exo- (3’ to 5’ exo minus). Paired end Index DNA adaptors (Agilent) with a single “T” base overhang at the 3’ end were ligated, and the resulting constructs were purified using AMPure SPRI beads from Agencourt (Beckman Coulter Genomics, Danvers, MA). The adapter-modified DNA fragments were enriched by 4 cycles of PCR using SureSelect forward and SureSelect Pre-Capture indexing reverse (Agilent) primers. The concentration and size distribution of the libraries was determined via Agilent Bioanalyzer DNA 1000 chip. Custom capture of 3.69 Mb was carried out using the Agilent Bravo liquid handler following the protocol for Agilent’s SureSelect XT. 750 ng of the prepped library was incubated with whole exon biotinylated RNA capture baits supplied in the kit for 24 hours at 65 °C. The captured DNA:RNA hybrids were recovered using Dynabeads MyOne Streptavidin T1 (Life Technologies, Carlsbad, CA). The DNA was eluted from the beads and purified using Ampure XP (Beckman Coulter Genomics). The purified capture products were then amplified using the SureSelect Post-Capture indexing forward and index PCR reverse primers (Agilent) for 12 cycles. Libraries were validated and quantified on the Agilent Bioanalyzer. Libraries were pooled at equimolar concentrations in batches of 96 samples and loaded onto paired end flow cells at concentrations of 7-8 pM to generate cluster densities of 600,000-800,000/mm2 following Illumina’s standard protocol using the Illumina cBot and HiSeq paired-end cluster kit version 3 (Illumina, San Diego, CA). The flow cells were sequenced as 101 X 2 paired end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HiSeq data collection version 1.4.8 software. Base-calling was performed using Illumina’s RTA version 1.12.4.2 (Illumina). Analysis We used Genesifter® software (PerkinElmer, Danvers, Massachusetts) to analyze the data. Within this program, raw reads from the sequencing above in fastq format were first trimmed to remove adaptor/primer sequences. Trimmed reads were then aligned using BWA against the genomic reference sequence for Homo sapiens (Build 37.2). The Homo sapiens (Build 37.2) reference sequence and annotation were pulled from the NCBI database (http://www.ncbi.nlm.nih.gov/). An additional alignment, post-processing set of tools were then used to do local realignment, duplicate marking, and score recalibration to generate a finale genomic aligned set of reads. Reads mapping to the genome were characterized as exon, intron, or intergenic (outside any annotated gene) using Build 37.2 for the Homo 4 sapiens reference. Nucleotide variants were called using GATK (Broad Institute, Cambridge, MA), which identified single nucleotide and small insertion/deletion (indel) events using default settings. PCR Validation We used both patient eosinophil (CD16-) and T-lymphocyte DNA for validation of somatic ETNK1 mutations in every case. Primers for amplification of ETNK1 exon 3 (ENST00000266517) were: ETNK1FP: 5’-AAGAAGATTCGGGAGA-3’, and ETNK1-RP: 5’-CAGCCAAAGAATCAATGC-3’. The 50µL reaction contained 20ng of DNA, and also included 5µL 10X reaction buffer (Roche, Indianapolis, Indiana), 1.5µL of a 25mM mixture of dNTPs, 2µL each of a 10mM concentration of both forward and reverse primers, 0.5µL of Taq polymerase (Roche), and dH20. Amplification included an initial denaturation at 94oC for 2 min, followed by: 35 cycles of a melt at 94oC for 30sec, annealing at 52oC for 45sec and extension at 72oC for 40sec, and ended with a final extension at 72oC for 3 min. Primers for screening ETNK2 exon 3 (ENST00000367202) were as follows: ETNK2-FP: 5’GGCCTGGAGGGTCCCTATCTGG-3’, ETNK2-RP: 5’-CCCGAGACCTAAGCCCTTAAAC-3’. Amplification for ETNK2 included the same master mix preparation as above and protocol as follows: denaturation at 94oC for 2 min, followed by: 40 cycles of 94oC for 20sec, 54oC for 15sec, 72oC for 45sec, with a final extension at 72oC for 1 min 30 sec. All products were visualized via 1.3% agarose gel and ETNK1 and ETNK2 mutational status confirmed by standard bi-directional Sanger sequencing. 5 Supplementary Figure 1. Nucleotide sequence conservation between species for ETNK1 gene in vicinity of region encoding for amino acid residues N244 and G245 6