EK Bio1 Ch 2 (cont..) Regulation of transcription In prokaryotes

EK Bio1 Ch 2 (cont..)
Regulation of transcription o In prokaryotes
mRNA typically includes several genes in a single transcript
Jacob-Monod model
Operon = operator + promoter + genes contributing to single prokaryotic mRNA o Genes outside of operon may code for activators/repressors
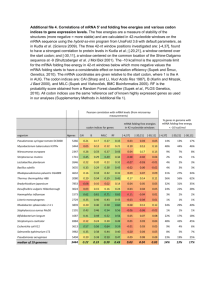
Lac operon in e. coli o Codes for enzymes that allow e. coli to import & metabolize lactose when glucose is nor present in sufficient quantities o Activated when glucose is scarce and lactose is present o Mechanism: (38-39)
Low glucose levels -> high cAMP levels
cAMP activates CAP
CAP binds to CAP site adjacent to promoter on lac operon
Positive control: CAP activates promotor
Gene repression: when lactose not present, lack repressor protein binds to operator, prevents expression of lac genes o In eukaryotes
mRNA includes only 1 gene per transcript
modification of RNA o post-transcriptional modifications offur in both eukaryotic and prokaryotic cells
eukaryotes: only in nucleus
bacterial genome does not contain introns o purpose of modifications:
help molecules that initiate translation to recognize the mRNA
protect mRNA from degradation
eliminate extraneous sequences of nucleotides from the transcript before translation
provide mechanism for variability in protein products produced by a single transcript o primary transcript = first nucleotide sequence arrived at through transcription
much longer than mRNA that will be translated into protein
portions of transcript (introns) are spliced out before exiting nucleus
exons are joined end-to-end
**introns stay IN nucleus, exons Exit nucleus to be translated
snRNPs = small nuclear ribonucleoproeins, act as ribozymes o spliceosome = complex of snRNP + associate proteins o 5’ end of eukaryotic transcription is capped via GTP before transcription is finished
5’ cap serves as attachment site in protein synthesis & as protection against enzyme degradation (via exonucleases)
3’ end has long series of adenine nucleotides (poly A trail) o alternative splicing in eukaryotic cells
allows cell to incorporate different coding sequences into mature mRNA
can create variety of mRNA molecules for translation from single DNA coding seq
human genome is 20,000 – 25,000 coding regions, but codes for >100,000 protein products
introns help determine possible splicing patterns & promote protein production
introns tend to be highly conserved between species
translation: nucleotide sequence of mRNA to amino acid seq of corresponding protein o triplet code: 3 nucleotides (codon) to code for 1 amino acid
degenerative… >1 series of 3 nucleotides can code for same AA
**Stop codons: UAA, UGA, UAG
**Start codon: AUG
Number of possible combinations of any three nucleotides: 4 3
= 64 o Types of RNA
mRNA = template that carries genetic code from nucleus to cytosol in form of codons
tRNA
two ends: o one contains anticodon, series of 3 nucleotides that binds to complementary codon sequence on
RNA o other carries AA that corresponds to codon, to be added to growing polypeptide chain
some flexibility in bonding at the third-bp position
wobble pairing o all translation uses ribosomes
small subunit + large subunit
prokaryotic: 30s + 50s = 70s
eukaryotic: 40s + 60s = 80s
the nucleoleus manufactures ribosomes (euk only) o mechanism: (p 43)
initiation
after post-transcriptional processing (euk), mRNA leaves nucleus though nuclear pores, enters cytosol
initiation factors help attach 5’ end to small ribosomal subunit
tRNA containing 5’-CAU-3’ gathers methionine and settles into p-site
this signals large subunit to join & form initiation complex o elongation
ribsosome slides down mRNA one strand at a time 5’->3’ while matching each codon to complimentary tRNA anticodon
corresponging AAs attached to tRNA are bound together into growing polypeptide
elongation requires energy
when tRNA attaches to P-site, new tRNA attaches to neighboring A site, peptide bond forms between the two via peptidyl transferase activity
tRNA with methionine moves to E-site, and then exits ribosome, tRNA carrying new dipeptide moces to p side, a site is open for next until stop codon reaches p o termination
when stop codon reaches A site, release factor proteins add to end of polypeptide chain, which frees it from tRNA and ribosome
ribosome breaks into subunits to be used later o polypeptide begins folding during translation, assisted by chaperons
post-translational modification o regulate gene expression by affecting which translational products become proteins o may add sugars, lipids, or phosphate groups, or cleave the polypeptide o final destination = related to location of translation o 20 AA seq called signal peptide near front of polypeptide is recognized by protein called RNA signal-recognition particle (SRP) that carries entire ribosome complex to receptor protein on ER
protein grows across membrane, then either released into lumen or remains partially attached to ER
signal peptide removed by enzyme
signal peptides may also be attached to polypeptides to target them to mitochondria, nucleus, or other organelles
DNA Replication: Mitosis o Semiconservative: each DNA copy contains one strand of original DNA o Mechanism governed by group of proteins called replisome
o Replication begins in middle of chromosome, at the origin of replication
Single origin on prok, multiple on euk.
Bidirectional process, two replisomes proceed in opposite directions