Ly br Sb + + +
advertisement
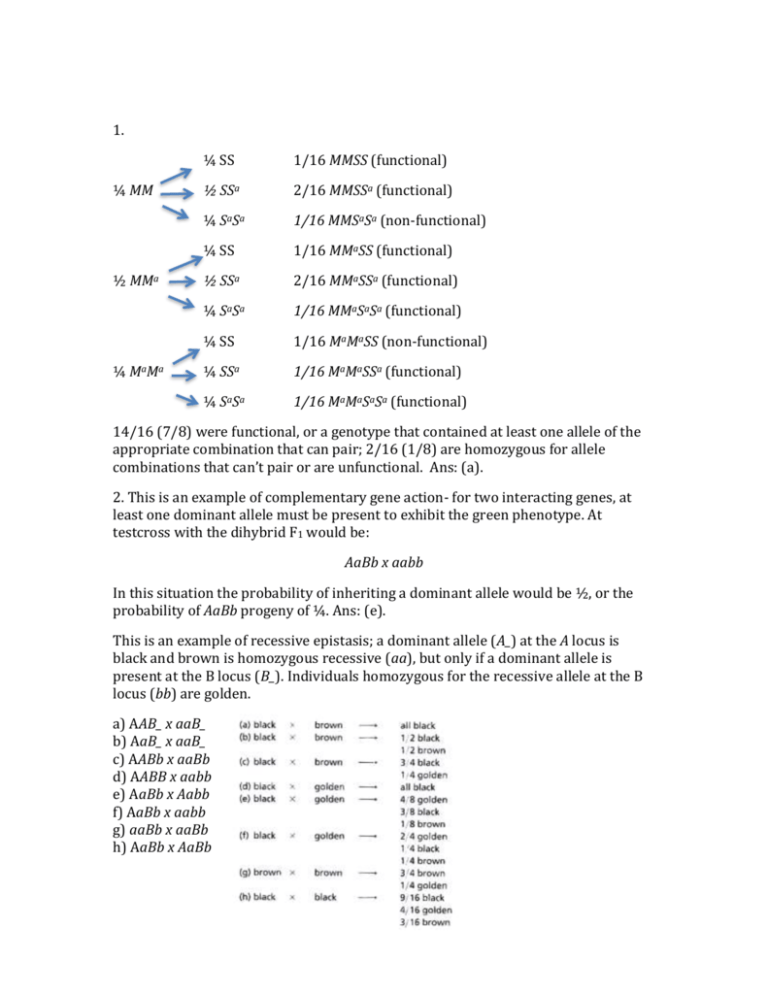
1. ¼ MM ½ MMa ¼ MaMa ¼ SS 1/16 MMSS (functional) ½ SSa 2/16 MMSSa (functional) ¼ SaSa 1/16 MMSaSa (non-functional) ¼ SS 1/16 MMaSS (functional) ½ SSa 2/16 MMaSSa (functional) ¼ SaSa 1/16 MMaSaSa (functional) ¼ SS 1/16 MaMaSS (non-functional) ¼ SSa 1/16 MaMaSSa (functional) ¼ SaSa 1/16 MaMaSaSa (functional) 14/16 (7/8) were functional, or a genotype that contained at least one allele of the appropriate combination that can pair; 2/16 (1/8) are homozygous for allele combinations that can’t pair or are unfunctional. Ans: (a). 2. This is an example of complementary gene action- for two interacting genes, at least one dominant allele must be present to exhibit the green phenotype. At testcross with the dihybrid F1 would be: AaBb x aabb In this situation the probability of inheriting a dominant allele would be ½, or the probability of AaBb progeny of ¼. Ans: (e). This is an example of recessive epistasis; a dominant allele (A_) at the A locus is black and brown is homozygous recessive (aa), but only if a dominant allele is present at the B locus (B_). Individuals homozygous for the recessive allele at the B locus (bb) are golden. a) AAB_ x aaB_ b) AaB_ x aaB_ c) AABb x aaBb d) AABB x aabb e) AaBb x Aabb f) AaBb x aabb g) aaBb x aaBb h) AaBb x AaBb 3. False, two gene loci are involved since we see a modified dihybrid (9:3:3:1) ratio in these crosses. 4. See above chart- the first parent is heterozygous at the A locus. Ans: (a) 5. Recessive epistasis: Ans: (d) 6. Ly br Sb + + + Ly------br interval: Ly + + 18 + br Sb 16 Ly + Sb 4 + br + 2 Total = 40/1000 = 4 map units br-------Sb interval: Ly br + 75 + + Sb 59 Ly + Sb 4 + br + 2 Total = 140/1000 = 14 map units 6. The distance from Lyra to Stubble is 18 map units. Ans: (e) 7. 0.04 x 0.14 x 1000 = 5.6 expected dcos. For this data set, 6 observed dcos… COC = 6/5.6 = 1.07 1-1.07 = -0.07 Ans: (a) True, no interference is seen in this data set…. The issue was raised in class that the Thompson book says that negative interference is possible- and it is. So I will count False as a correct answer here. HOWEVER, a three point test cross would not normally be a situation where you would see more crossovers than predicted (and here you really don’t- the numbers are virtually identical. Negative interference is rare and usually seen (in eurkaryotic organisms) in very special situations in fungi and yeast. 8. Are all three genes linked? If so, we would not expect independent assortment; but the A and B loci, and the C and B loci, appear to assort independently; i.e. a 1:1:1:1 ratio is observed among progeny in terms of gamete recovery: aB 214 + 30 ab 226 + 36 AB 26 + 218 Ab 222 + 28 cB 30 +218 cb 222 + 36 CB 214 + 26 Cb 28 + 226 But the A and C genes do not assort independently: Ac 218 + 222 = 440 AC 26 + 28 = 54 aC 226 + 214 = 440 A V ac 30 + 36 = 66 a c B b C b B X 54 + 66 = 120/1000 = 0.12 Distance = 12 m.u. Ans: (c) Tetrad Class 1 2 3 4 5 6 7 c+ c+ cd +d c+ cd c+ c+ cd cd c+ ++ ++ +d +d ++ ++ c+ cd cd cd +d +d ++ +d +d ++ ++ 1 17 41 1 5 3 1 9. Distance between c and centromere: 1+ 5 + 3 + 1 = 10 10 * 0.5/69(1/2) = 7.3 map units Ans: (a) 10. PD = 44; NPD = 2; so genes are linked… Distance between d and centromere: 22/69(1/2) = .0159 = 15.9 map units RF = (½ TT + 3NPD)/total = (23/2 + 6)/ 69 = 25.3 m.u. 7.3 + 15.9 = 23.3 m.u. Ans: (b)