A Study on the Characteristics of Linear Polymer Molecules
advertisement

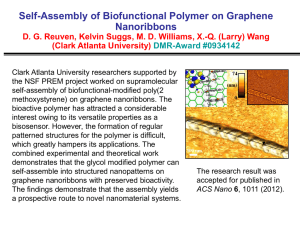
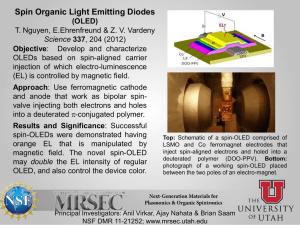
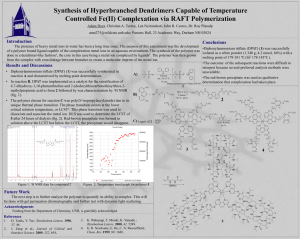
An Investigation on Characteristics of Linear Homopolymer Molecules Using Monte Carlo Simulation with Random Walk and Self-Avoiding Walk Models Idtisak Paopo, Pajaree Rittiyong, Siripon Anantawaraskul* Department of Chemical Engineering, Faculty of Engineering, Kasetsart University, 50 Phaholyothin Rd., Jatujak, Bangkok, Thailand, 10900 * Tel (662) 942-8555 ext 1231 Fax (662) 561-4621 E-mail fengsia@ku.ac.th ABSTRACT In this work, polymer characteristics (e.g., radius of gyration and end-to-end distance) of linear, monodisperse homopolymer molecules were investigated using Monte Carlo simulation with random walk (RW) and self-avoiding walk (SAW) models in two-dimension square lattice and three-dimension cubic lattice. The results show quantitative relationships between the number of steps (i.e., molecular weight) and the chain characteristics. The results from our simulation were validated with results from theory and data from literature for the limiting cases and a good agreement was obtained. The results from the present work will serve as a basis for further investigation of the characteristics of polymer molecules having various complex architectures of interests. KEYWORDS: Monte Carlo simulation, random walk (RW) model, self-avoiding walk (SAW) model, Size and shape of polymer molecule INTRODUCTION A molecular size is the basic characteristic of a polymer molecule. Difference in molecular sizes is the basis of the fractionation process in gel permeation chromatography (GPC), an important, widely used polymer characterization technique. In this technique, polymer molecules are fractionated by the hydrodynamic volume (reflected through the radius of gyration) to provide the information of molecular weight distribution (MWD). The knowledge of the relationship between molecular size and molecular weight is crucial and could help improve our understanding of this important characterization technique, especially in the case of polymers with complex chain architectures (e.g., branched, crosslinked, star, and comb polymers) [1-9]. These polymers with complex chain architectures have been found to be very useful and increasingly focused in both academia and industry. Therefore, understanding the characteristics of these polymers would be of interests. Two of the parameters that are commonly used for representing size and shape of polymer molecules are the mean square radius of gyration (<Rg2>) and mean square end-to-end distance (<Re2>). The radius of gyration is the distance of the segments from the center of gravity of the polymer coil and the end-to-end distance is simply the distance between both ends of the molecule. These parameters of each polymer molecule can be defined as follows [10]: Re (ωN ωo ) 2 2 R g 2 1 2(N step 1)2 (1) N (ω i ω j )2 (2) i,j 0 where Nstep is the number of steps (or segments) and ωi is a coordinate vector of structural unit i ([x, y] for 2D square lattice and [x, y, z] for 3D cubic lattice). In this work, we take a first step by investigating the relationship between molecular size (e.g., radius of gyration and end-to-end distance) and molecular weight in the case of linear, monodisperse homopolymers using Monte Carlo simulation technique together with random walk (RW) and self-avoiding walk (SAW) models in both 2 and 3 dimensions. The results from the present work will serve as a basis for further investigation of the characteristics of polymer molecules having various complex architectures. MONTE CARLO SIMULATION WITH RANDOM WALK (RW) AND SELF-AVOIDING WALK (SAW) MODELS For generating each polymer chain, Monte Carlo simulation technique was used with random walk (RW) or self-avoiding walk (SAW) models. Monte Carlo simulation is a widely used computational technique, especially for describing the statistical nature of polymer chains [1-4]. Details of this simulation approach with random sampling technique have been described in the literature [10]. This simulation approach is chosen in this study because of its main advantage over other methods (e.g., renormalization, exact-enumeration), as it allows us to probe information directly in the regime of fairly long polymer chain [10]. Especially with the advances in the computing technology, researchers now can easily perform some computation works, which are once considered too time-consuming, with a good resolution. Models of single polymer molecule Both RW and SAW models used in this study represent a polymer chain in different situations. In the RW model, a polymer chain is generated with the assumption that allows the chain to pass through the regions of space that are already occupied by other segments of the same chain. A polymer chain simulated using this model can represent the situation where a polymer molecule is surrounded by other chains of the same type. Because various interactions cancel out due to a collection of chains of the same type, a polymer chain in this situation behaves as if it could “pass through itself” [11]. In the SAW model, each polymer chain is generated with a condition that no segment is allowed to be in the same position. This model is well known for describing a polymer chain in a good solvent (excluded volume condition). In this work, we used SAW model with an algorithm called dimerization (algorithm based on the principle of “divide and conquer”) together with one called non-reversal random walks (NRRWs or memory-2 walks) [10]. Both RW and SAW models were investigated in 2D square lattice and 3D cubic lattice. Figure 1 shows examples of polymer chains with 100 steps obtained from different models and different lattices. In each model, linear polymers with the number of steps (Nstep) of 15-200 and the number of chains (Nchain) of 200,000 were studied to establish the quantitative relationships between Nstep and chain characteristics. 2D RW model 2D SAW model 6 4 0 2 0 -10 Y Y -2 -4 -20 -6 -8 -30 -10 -5 0 5 10 15 20 25 30 -6 -4 -2 0 2 X 4 6 8 10 X 3D SAW model 3D RW model 8 4 6 2 4 2 0 Z Z 0 -2 -2 -4 16 14 12 10 -6 -8 8 6 -6 4 6 8 6 4 4 2 2 0 Y -2 -4 X 8 -10 2 -8 6 4 0 2 -6 -8 -2 -2 0 0 -2 Y X -4 -4 -4 Figure 1 Examples of polymer molecules with Nstep= 100 generated from different models and different lattices. Figure 1 Examples of polymer molecules with Nstep = 100 generated from different models and different lattices. Model validations To validate our model, we compared results from our 2D-SAW model with the data from 2D-SAW model in the literature [12] in the case of Nstep = 15 and 20 (see Table 1) and a good agreement was obtained. This indicates that our algorithm is reliable can be used in a comprehensive investigation of long chain molecules. The comparisons between the results from other models (2D-RW, 3D-RW, and 3D-SAW) and the results from theory and literature [11-13] give a similar impressive agreement. Table 1 Comparison between the characteristic data obtained from the present work with Nchain = 200,000 and literature [12] Nstep 15 20 Present work 6.7842 10.2470 Radius of gyration Literature % Difference [12] 6.7843 0.001% 10.2477 0.01% Present work 47.2140 72.1034 End-to-end distance Literature % Difference [12] 47.2177 0.01% 72.0765 0.04% RESULTS AND DISCUSSION Effect of the number of chains (Nchain) It is well known that when using Monte Carlo simulation, a large amount of polymer chains has to be generated in order to obtain a good representation of all molecules. A small number of chains typically can not adequately represent the characteristics of the overall population and often lead to unreliable results. In order to determine the appropriate number of chains, a number of simulations was carried out with different number of chains in the case of Nstep = 15 and plotted in Figure 2. From Figure 2, it is obvious that the value representing characteristic of polymer molecule (mean square radius of gyration in this case) converges to a specific value, as Nchain increases. From these results, we chose the appropriate Nchain to be 200,000 because at this Nchain the standard deviation is already small and further increase in Nchain does not give a significant improvement of results. This Nchain will be used through out this work. 2 Mean square radius of gyration, <Rg > 7.6 7.4 7.2 7.0 6.8 6.6 6.4 6.2 6.0 1e+1 1e+2 1e+3 1e+4 1e+5 1e+6 1e+7 1e+8 Number of chains (Nchain) Figure 2 Effect of the number of chains on the mean square radius of gyration obtained using 2D SAW model with 15 steps. Effect of the number of steps (Nstep) The relationship between the molecular weight and polymer size is of interests in this study. Figure 3 illustrates relative sizes of polymer molecules with different numbers of steps obtained from the 2D SAW model. The quantitative effect of chain length (represented by Nstep) on polymer size (represented by mean square radius of gyration and mean square end-to-end distance) in case of 2D SAW model is shown in Table 2 and Figure 4. The results show that as the number of steps increases, both mean square radius of gyration and mean square end-to-end distance exponentially increase and the ratio between the mean square radius of gyration and mean square end-to-end distance converges to ~0.1403. This is in a very good agreement with the results (0.14026 ± 0.00007) reported earlier by Li et al. [13]. 30 30 Nstep= 20 20 Nstep= 50 20 10 10 0 0 -10 -10 -20 -20 -30 -30 -20 -10 0 10 20 30 -20 30 0 10 20 30 0 Nstep= 100 20 -20 0 -30 -10 -40 -20 -50 -20 -10 0 10 20 Nstep= 200 -10 10 -30 -30 -10 30 -60 -30 -20 -10 0 10 20 30 Figure 3 Relative sizes of polymer chains having different numbers of steps obtained using 2D SAW model Table 2 Effect of the number of steps on the characteristics of polymer chains (Nchain = 200,000) for 2D-SAW model <Rg2> 6.78427 10.24708 28.11626 51.17896 78.2744 109.3031 143.4247 Nstep 15 20 40 60 80 100 120 <Re2> 47.21401 72.10342 199.6772 364.7749 557.1227 779.1172 1,023.5282 <Rg2>/<Re2> 0.143692 0.142116 0.140809 0.140303 0.140498 0.140291 0.140128 1200 Radius of gyration End-to-end distance 1000 Distance 800 600 400 200 0 0 20 40 60 80 100 120 140 Number of steps (Nstep) Figure 4 Effect of the number of steps on the sizes of polymer chains A careful look at the results indicates that root of both mean square radius of gyration and mean square end-to-end distance is proportional to Nstep0.75 as showed by the linear relationship in Figure 5. We obtained the following relationships from our simulation results. Re 1 / 2 0.8839N step 3/ 4 Rg 1 / 2 0.3312N step 3/ 4 2 2 (3) (4) This again is in a good agreement with the results from theory [11], which indicates that Re 1 / 2 N step Rg 2 2 1/ 2 N step (5) (6) where 3 and d is the dimensionality of space (2 for square lattice and 3 for cubic lattice). d2 The results we obtained in Eq. (3) and (4) will be useful as a basis for comparison with polymer molecules with complex chain architectures in the further investigation. 35 Radius of gyration End-to-end distance 30 Distance 1/2 25 <Re2>1/2 = 0.8839 Nstep0.75 r2 = 0.9999 20 15 10 <Rg2>1/2 = 0.3312 Nstep0.75 5 r2 = 0.9998 0 0 10 20 30 40 Nstep0.75 Figure 5 Relationship between Nstep0.75 and square root of characteristic distances As mentioned before that RW and SAW models describe polymer molecules in different situations, the information about quantitative relationships between the number of steps and polymer characteristics for all the models is of important. In this article, we mainly show the results from 2D-SAW model to convey the key concept of our approach. The results of other models (2D-RW, 3D-RW, and 3D-SAW models) have also been validated and investigated in the same manner. Table 3 shows the summary of the final quantitative relationships we obtained for all investigated models. Table 3 Summary of quantitative relationships between the number of steps and polymer characteristics (<Rg2> or <Re2>) of all investigated models Model 2D-RW Relationship for radius of gyration Relationship for end-to-end distance Rg 0.1667N step R g 0.9832N step 3D-RW Rg 0.1683N step Rg 1.007N step 2 2 2 2 2D-SAW Rg 1 / 2 0.3312N step 3D-SAW Rg 1 / 2 0.4102N step 2 2 3/ 4 Re 1 / 2 0.8839N step 3/5 Re 1 / 2 1.0301N step 2 2 3/ 4 3/5 The results from Table 3 indicate that for RW models, the dimensionality (2D or 3D) does not affect the relationship between Nstep and polymer characteristics (Rg and Re). However, for SAW models, the dimensionality has a significant effect due to the self-avoiding or excluded volume effect. CONCLUSION In this study, we investigated polymer characteristics (e.g., radius of gyration and end-to-end distance) of linear polymer molecules using Monte Carlo simulation with random walk (RW) and self-avoiding walk (SAW) models in two and three dimensions. For the limiting cases, the present results from simulation were validated with results from theory and data from literature and a good agreement was obtained. From this investigation, we obtained quantitative relationships between the number of steps (i.e., molecular weight) and polymer characteristics (Rg or Re) for all the models. These relationships developed in this work can provide the information of the characteristics of long chain molecules, cases mainly disregarded in the past due to the long computational time required. This information will be useful as a basis for further research works on the characteristics of polymer molecules having various complex architectures, which are under investigation in our laboratory. ACKNOWLEDGEMENT The authors would like to thank the financial support for establishing computing facilities in polymer computation laboratory by petroleum and petrochemical technology consortium (ADB project). Reference [1] [2] [3] [4] [5] [6] [7] [8] Tobita, H. and Hamashima, N., ‘Monte Carlo Simulation of Size Exclusion Chromatography for Randomly Branched and Crosslinked Polymers’, Journal of Polymer Science: Part B: Polymer Physics, 38, pp 2009-2018, (2000). Tobita, H. and Saito S., ‘Size Exclusion Chromatography of Branched Polymers: Star and Comb Polymers’, Macromolecular Theory and Simulation, 8, pp 513-519, (1999). Tobita, H., ‘Random Sampling Technique to Predict the Molecular Weight Distribution in Free-Radical Polymerization That Involved Polyfunctional Chain Transfer Agents’, Macromolecules, 29, pp 693-704, (1996). Tobita, H. and Hamashima, N., ‘Monte Carlo Simulation of Size Exclusion Chromatography for Branched Polymers Formed through Free-Radical Polymerization with Chain Transfer to Polymer’ Macromolecular Theory and Simulation, 9, pp 453-462, (2000). Zhou, Z. and Yan, D., ‘Mean-Square Radius of Gyration of Polymer Chains’, Macromolecular Theory and Simulation, 6, pp 597-611, (1997). Zifferer, G., ‘Distribution of the Shape Asymmetry of Linear and Star-Branched Random Walks’, Macromolecular Theory and Simulation, 6, pp 805-824, (1997). Zifferer, G., ‘Monte Carlo Simulation Studies of the Size and Shape of Linear and StarBranched Polymers Embedded in the Tetrahedral Lattice’, Macromolecular Theory and Simulation, 8, pp 433-462, (1999). Zifferer, G., ‘Shape Asymmetry of Star-Branched Random Walks and Nonreversal Random Walks’, Macromolecular Theory and Simulation, 6, pp 381-392, (1997). [9] [10] [11] [12] [13] Zimm, B.H. and Stockmayer, W.H., ‘The Dimensions of Chain Molecules Containing Branches and Rings’, The Journal of Chemical Physics, 17(12), pp 1301-1314, (1945). Sokal A.D., ‘Monte Carlo Methods for the Self-Avoiding Walk’, in: Monte Carlo and Molecular Dynamics Simulations in Polymer Sciences, Binder, K., Eds., Oxford University Press, Oxford, (1995). Painter, P.C. and Coleman, M.M., Fundamentals of Polymer Science. Technomic Publishing Company, Inc., Pennsylvania, (1997). Madras, N. and Sokal, A.D., ‘The Pivot Algorithm: A Highly Efficient Monte Carlo Method for the Self-Avoiding Walk’, Journal of Statistical Physics, 50, pp 109-186, (1988). Li, B., Madras, N. and Sokal, A.D., ‘Critical Exponents, Hyperscaling, and Universal Amplitude Ratios for Two- and Three-Dimensional Self-Avoiding Walks’, Journal of Statistical Physics, 80, pp 661-754, (1995).