Materials and Methods
advertisement

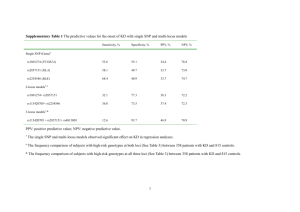
1 Dyskinesia-associated glutamate receptor genotypes: Materials and Methods Personal contribution SI and AL instigated, designed, coordinated and supervised the study; AL had the idea, estimated the genes and financed the study with lecture fees; PP and MF designed and performed the statistical analysis and contributed to writing the paper; AAH wrote the study protocol and picked the tSNPs; ER and OF monitored the study, collected clinical data and isolated DNA. ER genotyped the samples and recorded all data in an excel data base; IZ, NG, VS, VA, AS examined the patients and collected patient data. VA and AS supervised the clinical work. SI, AL, JB and BW supervised the technical work. AL wrote and revised the manuscript. SI, PP, MF, AAH, JB, and BW commented on the original and revised manuscript. Patients We examined a total of 574 patients. The protocol was approved by the local Ethical Committee and all subjects gave informed consent after a complete explanation of the study. Of the 574 participants, 431 were from three psychiatric hospitals in Tomsk (two centers), Kemerovo, and Chita oblasts in Siberia. These patients demonstrated schizophrenia or schizotypical disorders according to criteria from the International Statistical Classification of Diseases and Related Health Problems, 10th Revision (ICD-10). All these patients were on long-term treatment with antipsychotic drugs. We excluded non-Caucasians, patients with addiction problems, and those with neurological or organic disorders. Patients were assessed for the presence or absence of dyskinesia by adequately trained medical doctors under the direct supervision of the experienced local primary investigator, according to the abnormal involuntary movement scale (AIMS).15 No formal joint training sessions were organized, which might be considered as a possible limitation of the present study. In addition, a blood sample was taken for DNA isolation and genotyping. Inclusion criteria were no current clozapine treatment (clozapine suppresses TD) and high quality DNA samples. We also examined 143 patients with spontaneous extra-pyramidal disorders (101 had been diagnosed with Parkinson’s disease) from the neurology department of the Siberian State Medical University of Tomsk. These patients, who 2 were originally intended to serve as a reference group, had not been treated with dopamine antagonists (antipsychotic or antiemetic drugs) for at least three years. Other inclusion criteria were Caucasian race, no addictions, no current clozapine treatment, no organic disorders, and a high quality DNA sample. Patients were asked to refrain from alcohol and/or drug intake for 12 h prior to assessment. On the day of assessment, complete data were collected on medication use (name and dose of all medicines used). Moreover, an examination was performed to ensure that no clinically-relevant withdrawal symptoms were present. Demographic, clinical, and medication data were extracted from patient medical files (Table 1). For comparisons, antipsychotic medication doses were converted into chlorpromazine equivalents.14 The AIMS scores were transformed into a binary form (presence or absence of dyskinesia) with Schooler and Kane’s criteria.16 Orofacial (AIMS items 1-4), limb-truncal (AIMS items 5-7), and ‘all locations’ (AIMS 1-7) dyskinesias were analyzed separately. The sum of AIMS items in an individual was used as a proxy for the severity of dyskinesia. DNA analysis We analyzed a panel of 48 tag SNPs in 8 genes. These included 15 SNPs that covered the GRIN2A gene (rs9989388, rs7190619, rs8049651, rs7196095, rs9921541, rs7192557, rs9788936, rs7206256, rs11866328, rs4782039, rs1345423, rs11644461, rs11646587, rs8057394, rs1650420) and 9 that covered the GRIN2B gene (rs12300851, rs220599, rs7313149, rs12827536, rs10772715, rs10845838, rs1805481, rs2192970, rs2300242). Moreover, 10 SNPs covered the DRD3 gene (rs167770, rs167771, rs324035, rs963468, rs1800828, rs2134655, rs3773678, rs7633291, rs9817063, rs11721264), 1 SNP the DRD4 gene (rs3758563), 5 SNPs the HTR2C gene (rs6318c, rs569959, rs1801412, rs3813929, rs4911871), 1 SNP the HSPG2 gene (rs2270697), 1 SNP the MnSOD gene (rs4880), and 1 SNP the NQO1 gene (rs1800566). Statistical analysis In order to be more certain about the validity of the results, we used two statistical approaches to analyze the data. All calculations were performed in the R statistical environment with the SNPassoc 19 3 and LogicReg18 package and basic R functions. Method 1: Logistic regression and log-linear regression First, allele prevalence was calculated separately in patients with psychiatric disorders and in patients with neurologic disorders to define the percentage of missing genotypes. The Hardy-Weinberg equilibrium test was applied with Fisher’s exact test to groups with and without dyskinesia. This showed that one SNP, rs12858300, was monomorphic; therefore, it was removed from further analysis. To analyze associations between the SNPs and the phenotypes, we used logistic regression for binary response traits and log-linear regression for continuous traits. The following genetic models were tested: 1. Co-dominant; both alleles of a SNP influenced the phenotype 2. Dominant; rare allele homo- and heterozygotes were tested against common allele homozygotes. 3. Recessive; common allele homo- and heterozygotes were tested against rare allele homozygotes. 4. Over-dominant; heterozygotes were tested against both homozygote alleles. 5. Log-additive; a trend test for the genotypes, similar to the allele model, but comparisons were among subjects (N) instead of chromosomes (2N). The test and estimates were based on a logistic regression model that coded the genotypes as 0, 1, or 2 to reflect the number of minor alleles. The statistical significance of a SNP was established with a likelihood-ratio test that compared the effect of a polymorphism with the null model (only including the intercept). The Akaike information criterion was applied to identify the model that best fit the data. SNP effects were quantified with the odds ratio (OR) and 95% confidence intervals. In the result table, an OR for a log-additive model corresponds to an association between a rare allele and the presence of dyskinesia. The ORs for other models correspond to associations between the presence of dyskinesia and rare allele homo- and heterozygotes (Dominant), common allele homo- or heterozygotes (Recessive), or heterozygotes 4 (Overdominant) Dyskinesia was considered a qualitative trait (yes/no), but the degree of dyskinesia expression was analyzed as a quantitative trait. To avoid 0 values, 1 was added to the dyskinesia expression values, and then they were log2-transformed to obtain a log-normal distribution. There were many confounding factors in patients with schizophrenia, including gender, age, duration of disease, use of anticholinergic medicine, dose of anticholinergic medicine, and total chlorpromazine equivalents on the day of assessment. Only gender and total chlorpromazine equivalents were determined to be insignificant. The genetic models were adjusted for all the significant confounding factors. In contrast, for data from patients with neurological disorders, the genetic models were only adjusted for age. Method 2: Permutation tests and logic regression In order to assess the level of signal information in the two datasets and identify possible SNP-SNP interactions of higher order, logic regressions were fitted to the data.18 Logic regression is a methodology developed primarily for identifying associations between binary explanatory covariates, and their interactions, with a binary or continuous outcome of interest.18 Logic regression requires that the covariates of interest are added in a binary fashion in the model. For this reason, we coded the SNPs of interest with two binary dummy covariates S1 and S2, as suggested by Schwender et al.:26 “S1: At least one of the bases explaining the SNP is the less frequent variant” “S2: Both bases explaining the SNP are the less frequent variant”. When missing values for a SNP comprised more than 10% of the sample, they were excluded from the analysis. Furthermore, when the DNA of a participant displayed more than 10 missing SNPs, it was excluded. The remaining missing values were imputed by random sampling from the marginal distribution of the proportions of alleles in the patient population.26. After imputation, we removed SNPs that had one absent variant. Permutation tests were used to identify the presence of any signal from the data. Cross validation methods were applied to assess the goodness of fit of the suggested model. Finally, a Monte Carlo 5 logic regression approach was used to identify the SNPs with the highest probability of being associated with the phenotype.19 All logic regression models were corrected for age and duration of disease. Models for patients with neurological disorders were also corrected for levodopa use. Additional reference 26. Schwender H, Ickstadt K. Identification of SNP interactions using logic regression. Biostatistics 2008; 9: 187-98.