WHAT IS AN E
advertisement

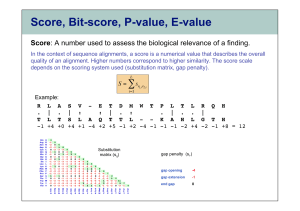
WHAT IS AN E-VALUE? A BLASTn search returns hits, sequences that produce “significant” alignments to the query sequence. The significance of a hit is measured by its E-value, or expect value. Each alignment has a bit score (S), which is a measure of similarity between the hit and the query; it is given in the column next to the E-value. The E-value of a hit is the number of alignments with bit score S that you expect to find by chance (i.e., with no evolutionary explanation). Biologically significant hits will tend to have E-values much less than 1.0. An E-value near 1.0, or even substantially larger than 1.0, does not necessarily mean that the corresponding hit is biologically irrelevant; however, the larger the E-value, the greater the chance that the similarity between the hit and the query is due to mere coincidence. E-values are calculated from the following three factors: 1. The bit score. Since a larger bit score is less likely to be obtained by chance than is a smaller bit score, larger bit scores correspond to smaller E-values. 2. Length of the query. Since a particular bit score is more easily obtained by chance with a longer query than with a shorter query, longer queries correspond to larger E-values. 3. Size of the database. Since a larger database makes a particular bit score more easily obtained by chance, a larger database results in larger E-values. To see how E-values are calculated, perform a "Compare Two Sequences" search with the following sequences: acaagctcagaatttaatttgtgcgaagctcagcattaatcgtacttcatatagccctg and acaagctcagaatttaatatgtgcgttgctcagacattaatcacttcatatagccctg Copy the alignment and relevant data into your answer sheet. Above each alignment in the BLASTn report is the associated bit score (S) and, in parentheses after the bit score, the raw score (R). For example, the first hit (E-value near 0.007) has S = 62.2 bits and R = 32. E-values are computed directly from bit scores, which are in turn computed directly from raw scores. Raw Scores The raw score is calculated by counting the number of identities, mismatches, gaps, and “-” characters in the alignment and setting R = aI + bX - cO - dG, where: I is the number of identities in the alignment, and a is the reward for each identity X is the number of mismatched nucleotides, and b is the “reward” for each mismatch O is the number of gaps, and c is the penalty for opening a gap G is the total number of “-” characters, and d is the penalty for each “-” in the gap The values of the parameters a, b, c, and d are printed at the bottom of the BLASTn report. The default values are a = 1, b = -3, c = 5, and d = 2; these values can be changed on the “Other advanced” line. For example, in our first alignment, there are 54 identities, 3 mismatches, 3 gaps, and 3 “-” characters. Calculate R by putting these values into the equation above. Do you get the same number as reported (in parenthesis beside the “score”? Bit Scores The bit score is obtained from the raw score using the equation where and K (also found somewhere on the Blast site—I just can't find everything anymore!) are normalizing parameters. Assuming =1.37 and K=0.711, and using the R value of 32, calculate the Bit score. As a result of the normalizing process, bit scores and E-values are independent of the scoring system, allowing those calculated with particular rewards and penalties to be compared directly to those calculated with different rewards and penalties. E-values Finally, the E-value is calculated as E=mn2-S, where m is the effective length of the query, and n is the effective length (total number of bases) of the database. (Effective lengths are adjusted from the actual lengths to account for the fact that an alignment cannot start near the end of the sequence, the so-called edge effect.) In our example, m=34 (19 nucleotides fewer than the 53 bases submitted) and n= 3,743,317. Plugging these numbers into the E-value equation gives E=0.007, as shown in the BLASTn report. Why is the E value different when you put only a smaller number of bases into the search, even though the bases are the same as some included in a larger sequence? Test another sequence, as follows (you can copy numbers—the program knows not to consider them): TTAACACGTGTTGTTGGGAGTTCGAACCTCCCCCTTCCAGCCATCTCTTCTGTTGGTGCA TAGCTTAATGGTAAAGCCCTGGTCTCCAAAACCAGTCACCTAGGTTCGATTCCTGGTGCA TCAGCCAAATCTGTCTAAAATAGTCCACCATGACAGATTTCCTCTTTGATCCCCACGCAC ATGTTAGGTTCGACAAGATCGATCTCGTACCAGAGGCTCAAAATCCTCGGTTCAAGGAGT TGATCGAAAATACCAACCTGAACTACAAATACGATCCGAATGCTCCTCTCATCGTTGACA CGACGAAGTACGGTCCTCGTTCATTCTACATGAACAATAAGAAGATCAAACGTACTGGCG TCGAAATCAACATGACTGATGAGATGGTGGAGGAGCTCTACAAGTGCTCGACTGATGTCT TGTACTTTGCTGAGCGATATTATTACATTCGTACTCTTGACCACGGCAAGATCAAGATCC CTCTCCGTGACTATCAGAAATTCTGGCTCCGCATTTACGAGGTCCCTGAGATCCGTAACC GCGTGTGGCTCGCTTGTCGTCAGTCAGCCAAGTCCACGACACTGACCGTTGAGATCATGC ATCGAATGCTCTTCAATGAGGATTTCGAATACGTCATCCTGGCCAACAAGGGTAACACGG CTCGTGAAATCTTCTCGCGTGTCCGTATGGCATATGAACAGCTTCCTCTCTGGATGCAGA TCGGCGTGACCGAATGGAACAAGGGTTCTGTGAAGCTCGAGAACGATTCCCGTGTCTTCG CTGCTGCTTCTGGTTCTGACTCTGTCCGCGGTTTCTCTCCCAACGAAGTTCTCCTCGACG AAGCGGCATTCGTTCGTAATGATGAAGAGTTCATGGCCTCCGTGTTCCCGACCATCTCGT CTGGTTCTAAGTCTCGTCTGACCCAGATCTCCACGCCGAATGGCCCTCGTGGGATCTTCC ACCGAGATTACACTCGTGCGACCAAGGGTCTCAACAACTACTTCTCCTACAAGGTTCCAT GGCACTTTGTTCCTGGCCGTGATGAGGAATGGAAGAAGAAGCAGATCGAAGATACCTCGC TCATGCAGTTCAAGCAGGAGCAGGATTGTGACTTCATGGGCACTACCGACGGCCTCATCG ATGGTATGGTTCTCGAGAGCATCCAGGACAATATTCAGGCCCCAATTCTTGTCAATGATG AGGAACTGTCGGATCCAGCGTATGCAATCTTCGAACTCCCGAAGGAAGGACACTCGTATA TCGTCACTGCCGACACCGGTGAAGGTAAAGGCAAGGACTCGAGCACATTCACCGTGTTTG ACGTTTCGACTCGTCCGTTCGTCCAGGTTGCTTCCTACAAATCTAACCAGGTTTCCCAGC TGATCTTCCCAAACAAGCTGGTCAAGATCGCAGAGACATACAATAATGCTCTCTTGATCC CAGAAAGAAATAACACATCCGGCGGCACTGTCTGCTATAAGGTCTATTATGAACTCGAAT ACCCGAACGTCTTCCTCCAGGGCGATGGCGAAATGGACATCGGCGTCCATGTCTCTCATG CTGTCAGATCCCTCGGTGTCAATACCCTCCGTGGCCTCGTTGAAAAGGGCGGTCTCATTA TCCGAGATGAACGGACCTTCAAGGAACTAGCGAACTTCCGTCTCCAGAAGAACGGGAAAT ATGCTGCTCCAGAAGGTGAACATGATGATATGGTCCAGAACCTTTGGATATTCTCGTGGT ATACGGCTGGTGACGAATTCGAAGAAGCGATGAAGGAAAATATCTACAATGACCTCTATC GTGAAGAGCTCCAGAGCATCGAGAACCTGAAGGTCACCTCTGCGAACGACGCTTATGACC CGTATAATGTGAAACCAGTGGCAGGAAAGGCGGCCTCTGCGTTTTGGTAAGAAAGGACAA ATGATGGCATATGATCTATTCTCGGGTACCGTGGCCAATATCCCGGACCTGTCCACTCTC AAAGAGAAGACGAGAGAGCTTCTCCAGATCCCTGTGTCGGCTGGCCTGGATATCACTCGC TGGCTGAACGCACCTTCTCTCAATATCCCGGACCCGTTCAAGGAGTTCAATTTCAAATTC AAGGGCGGTCGAGTATCGTTCGAGGAATATGAAAGGAATTTTCTTCCGAAAGTTCCTAAC ATTCTCCAGTCGGCCAATCCGATCGGGTTCCAGGATGTCATCAGTGTCTTGGAGACGTTG Perform a BLASTn analysis (NOT MEGABLAST). Note the color coding of the matches. What is the best match? Now choose BLASTx, which compares translated nucleotide sequences to protein sequences. What is the best match for this search? Why are the results different?