DealingWithStatisticalUncertaintyPractical
advertisement

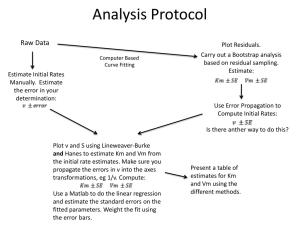
Dealing With Statistical Uncertainty – Practical Richard Mott Excercises This exercise explores properties of the Normal and T distributions. You will learn how to use some of the basic R functions associated with these distributions. You will also write a small R program to perform a permutation test and a bootstrap confidence interval. Remember that R will provide help on any function func() by typing the command ?func. You can get help on all the functions associated with the Normal and T distributions by typing ?Normal and ?TDist a. Graph the density functions of the standard Normal distribution N(0,1) and the T distributions on 1, 2, 3, 4, 5, and 10 degrees of freedom (df), between the limits x=-3 and x=+3, on the same plot. At what df does the T distribution become similar to the Normal? Hint: To do this, first generate a dense sequence of x values with the command x <- seq( -3, 3, 0.01) Then use this as input to the functions dnorm() and dt() eg to get the density for T10 use the command t10 <- dt(x,10) To plot more than one graph on the same plot, use plot() for the first plot and lines() for the rest. You can specify different colours use the col=”colour” parameter in lines()]. b. For the Normal distribution with mean 2 and variance 4, (i) what value of x satisfies Prob(X>x) = 0.05 (hint: use the function qnorm() ) (ii) what is Prob( X< -1) ? c. Generate a random sample of 10 observations from the above Normal N(2,2) distribution. (i) Calculate the sample mean and variance. (ii) Using the T distribution, calculate the 95% confidence interval for the sample mean (what df do you use?). (iii) Using a one-sample T test, test the null hypothesis Ho: =2.5. (iv) Now generate random samples of 100 and 10000 observations (save these samples into variables with different names eg samp10, samp100, samp10000, as you will re-use them later), and compute the 95% CIs again. What do you notice about the change in width? Repeat your tests of Ho: =2.5. d. Generate two samples of 20 observations, one from N(0,1) and the other from N(0.5,1). (i) Test if their means are different using the two-sample T-test and the Wilcoxon rank sum test. (ii) Add an outlier, by picking a data point at random and setting it to an untypical value, such as 5 or- 10. Now repeat your T and Wilcoxon tests. e. Download the Biochemistry.txt data set and read it into a data frame using read.table().This data set contains biomarker measurements on about 2000 mice. Check you have read in the file correctly by printing the column names (with names()) and the first few lines of the table (with head()). Test if there are GENDER effects present for Biochem.HDL using the formula interface to t.test(). The formula you will need is Biochem.HDL ~ GENDER. f. Perform power calculations using the function power.t.test(). (i) What sample size is required for a two-sample T test (equal variance=1, and equal numbers of observations in each group) to detect a difference between Ho: =2.5 and H1: =4.5 at =0.01 and =0.9. (ii) Using this number of observations, what is the power if H1: =4.0? g. Perform a permutation test on the two-sample data set in (d) above. You will need to write an R program to do this. Hint: First convert the data into a form suitable for permutation. The simplest way is to concatenate the two samples into a single vector, and create a second vector of the same length with 1’s and 2’s in it corresponding to the two groups. Then write a function that will accept these vectors as arguments and compute the difference between the means of the two groups. Now write a loop which will permute either vector (using the sample(replace=FALSE) function) 1000 times and call the function you wrote to compute the difference between the permuted groups, and increment a counter if the difference exceeds the real difference in the unpermuted data. Finally print out the permutation p-value. Compare it to that from the T test. h. Now modify your code to compute the bootstrap 95% confidence interval of the difference in means between the two groups. To make a bootstrap sample, use sample(replace-TRUE). Note that you must not permute the data. You will need to record the difference in means of the two groups at each bootstrap iteration in a vector. Then, at the end, sort the vector to find the 95% CI (this corresponds to finding the bottom 2.5% and top 2.5%) i. This exercise teaches you about simulation, in the context of a contingency table analysis. You will simulate data from a simple one-dimensional contingency table under the null hypothesis, compute the log-likelihood ratio and compare it to the asymptotic chi-squared distribution. (If you get stuck look at the function contingency.table() in examples.R) i. Simulate N=200 observations from a multinomial distribution with K=10 classes under the null hypothesis that all class are equally likely. Hint: type ?Multinomial to find out how to do this. ii. Compute the log-likelihood ratio L = 2 O log(O/E) iii. Repeat this say 10000 times. iv. Compare the distribution of L to the chi-square on K-1 df, eq with a qqplot v. Also, compare the distribution of the chi-square statistic (O-E)2 /E vi. What happens to the aymptotics when the sample size N is smaller (you will have to make your code handle zero counts)?