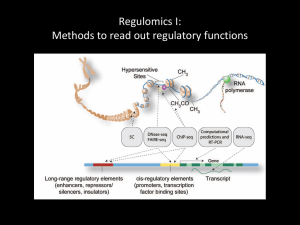
ChIP-Seq - the long version
advertisement

Analysis of ChIP-Seq Data Biological Sequence Analysis BNFO 691/602 Spring 2014 Mark Reimers What Are the Questions? • Where are histone modifications? • Where do TFs bind to DNA? • Where do miRNAs or RNABPs bind to 3’ UTRs? • How different is binding between samples? Why ChIP-Seq? • ChIP-Seq is ideal (and is now the standard method) for mapping locations where regulatory proteins bind on DNA – Typically ‘only’ 2,000 - 20,000 active binding sites with footprint ~200-400 base pairs • Similarly ChIP-Seq is fairly efficient for mapping uncommon histone modifications and for RNA Polymerase occupancy , because the genomic regions occupied are very narrow Chromatin Immuno-Precipitation Chromatin ImmunoPrecipitation (ChIP) is a method for selecting fragments from DNA near specific proteins or specific histone modifications From Massie, EMBO Reports, 2008 Chromatin Immuno-precipitation • Proteins are cross-linked to DNA by formaldehyde or by UV light • NB proteins are even more linked to each other than to DNA • DNA is fragmented • Antibodies are introduced • NB cross-linking may disrupt epitopes • Antibodies are pulled out (often on magnetic beads) • DNA is released and sequenced CLIP-Seq – A Related Assay • Cross-linking immuno-precipitation (CLIP)Seq is used to map locations of RNAbinding proteins on mRNA • Even miRNA binding can be mapped indirectly by CLIP-Seq with antibodies raised to Argonaute – an miRNA accessory protein What ChIP-Seq Data Look Like From Rozowsky et al, Nature Biotech 2009 The Value of Controls: ChIP vs. Control Reads Red dots are windows containing ChIP peaks and black dots are windows containing control peaks used for FDR calculation NB. Non-specific enrichment depends on protocol Need controls for every batch run Goals of Analysis 1. Identify genomic regions - ‘peaks’ – where TF binds or histones are modified 2. Quantify and compare levels of binding or histone modification between samples 3. Characterize the relationships among chromatin state and gene expression or splicing General Characteristics of ChIP-Seq Data • Fragments are quite large relative to binding sites of TFs • ChIP-exo (ChIP followed by exonuclease treatment) can trim reads to within a smaller number of bases • Histone modifications cover broader regions of DNA than TFs • Histone modification measures often undulate following well-positioned nucleosomes ChIP Reads Pile Up in ‘Peaks’ at TF Binding Sites on Alternate Strands ChIP-Seq for Transcription Factors • Typically several thousand distinct peaks across the genome • Not clear how many of lower peaks represent low-affinity binding sites From Rozowsky et al, Nature Biotech 2009 ChIP-Seq for Polymerase • Fine mapping of Pol2 occupancy shows peaks at 5’ and 3’ ends From Rahl et al Cell 2010 ChIP-Seq Histone Modifications • Many histone modifications are over longer stretches rather than peaks • May have different profiles • Not clear how to compare Issues in Analysis of ChIP-Seq Data • Many false positive peaks – How to use controls in data analysis – How to count reads starting at same locus • What are appropriate controls? – Naked DNA, untreated chromatin, IgG • Some DNA regions are not uniquely identifiable – ‘mappability’ • How to compare different samples? – Overlap between peak-finding algorithm results are often poor Mapability Issues • Many TFBS and histone modifications lie in low-complexity or repeat regions of DNA • With short reads (under 75 bp), with some errors, it may not be possible to uniquely identify (map) the locus of origin of a read • UCSC provides a set of mapability tracks – Select Mapping and Sequencing Tracks – Select Mapability – 35, 40, 50 & 70-mer mapability (some with different error allowances) END for Seq Analysis ChIP-Seq for Histone Modifications • Various histone modifications characterize different regulatory states Exon Peaks Intronic Peaks Intergenic H3K4me3 Peaks • Peak of H3K4me3 in region not annotated by RefSeq • Corresponds to unknown TAR in cerebellum • (annotated by Aceview) H3K4me3 vs Gene expression • 91.5% of expressed genes have H3K4me3 peaks H3K4me3 peaks at TSS peaks within 1 kb of the TSS CLIP-Seq for RNA Binding Proteins Thomson D W et al. Nucl. Acids Res. 2011;39:6845-6853 © The Author(s) 2011. Published by Oxford University Press. The ENCODE Project • Comprehensive characterization of chromatin state and locations of some TFs and other DNA-binding proteins (e.g. Pol2) across various conditions in human cell lines and in mouse tissues • So far no normal human tissues, which likely have rather different epigenetic marks Demo: ENCODE Data at UCSC ChIP-Seq Demo Peak Calling Goals of Peak-Calling • Identify discrete locations where a particular protein binds • Often applied more generally (and IMHO poorly) to identification of short regions with a particular histone modification Issues in Peak-Calling • Background of random genomic reads is not uniform – Affected by CG content and other factors • Most good algorithms try to estimate a local background • Local background has peaks too! Peak-Finding - Simple • Extend tags; sum overlaps at each base • Find center of each discrete cluster • Issues: – Are clusters discrete? – How much to extend? • Fragment size unclear Peak Finding – Better • Tags starting on opposite strands are likely to start at opposite ends of precipitated fragments • Identifying the cross-over point leads to better accuracy Issue: How to Identify a Cluster • Background varies – often related to local CG content and chromatin state • Need statistical test for excess counts above local background • Usually done by binning counts into 1kb bins across genome Control Reads Show Peaks Also From Rozowsky et al Nature Biotech 2009 Cause of Variation in Read Density • In study of FoxA1 binding, even control reads enriched near FoxA1 binding site! • Probably due to open chromatin near FoxA1 binding site Density of Control Channel reads around FoxA1 site Courtesy Shirley Liu Peak Finding by MACS • Smart peak imputation estimate – Uses read directions – Empirical estimate of fragment length • Local frequency estimate – Using control, if available – Using wide estimate, otherwise – Not using sequence MACS Workflow • Key innovation is to estimate fragment length empirically • If no control sample MACS estimates background from median of ChIP bin counts – No use of CG content The Value of Controls: ChIP vs. Control Reads Red dots are windows containing ChIP peaks and black dots are windows containing control peaks used for FDR calculation Issue: Fragment Lengths • Puzzle: Fragments from sonication expected to be between 200 – 500 bp • Empirically estimated fragment size ~ 100 • Shirley Liu’s explanation: preferential fragmentation near TF Peak Calling Demo Quantitative Comparison of ChIP Data