Epigenetics
advertisement

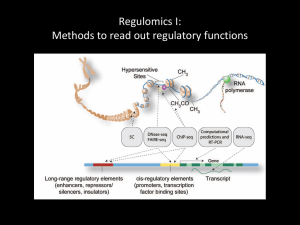
Regulomics II: Epigenetics and the histone code Jim Noonan GENE760 Biochemical indicators of regulatory function 1. TF binding 2. Histone modification 3. Chromatin modifiers & coactivators 4. DNA looping factors • H3K27ac p300 • H3K4me3 MLL cohesin Chromatin structure nucleosome H2A H3 H2B H4 H3 tail Luger, Richmond & colleagues 1997 Chromatin structure Zhou et al. Nat Rev Genet 12:7 (2011) Covalent modifications on histones me = methylation ac = acetylation ph = phosphorylation ub = ubiquitination H3 K27 ac Histone Residue Modification From Bannister and Kouzarides (2011) Cell Res 21:381 Covalent modifications on histones me = methylation ac = acetylation ph = phosphorylation ub = ubiquitination H3 K27 me3 Histone Residue Modification From Bannister and Kouzarides (2011) Cell Res 21:381 Histone modifications correspond to functions in the genome Zhou et al. Nat Rev Genet 12:7 (2011) Histone modifications correspond to functions in the genome Zhou et al. Nat Rev Genet 12:7 (2011) Histone modifications and chromatin accessibility H3K4me3 H3K4me3 + H3K27ac At promoters of developmental regulator genes in ES cells H3K27me3 Zhou et al. Nat Rev Genet 12:7 (2011) Writers, readers and erasers of histone modifications Tollervey and Lunyak (2012) Epigenetics 7:823 Ram et al., Cell 147:1628 (2011) Writers, readers and erasers of histone modifications MLL p300 PRC2 KDM6B From Bannister and Kouzarides (2011) Cell Res 21:381 Chromatin readers mediate functional outputs Musselman et al. (2012) Nat Struct Mol Biol 19:1218 Chi et al. (2010) Nat Rev Cancer 10:457 There are many chromatin regulators Ram et al., Cell 147:1628 (2011) Histone variants • Low abundance—limited copy number • Primary sequence variation • Specific functions – CENP-A: centromere – H3.3: transcription – H2A.Z: euchromatin/ heterochromatin boundary, TSS – H2A.X: genome integrity and high order structure From Tollervey and Lunyak (2012) Epigenetics 7:823 Genome browser interlude Using chromatin profiling to characterize biological systems Comparing chromatin states across cell or tissue types • Combinatorial analyses of histone modifications • Global transcriptome profiling • TF binding/motif analyses Identify cell- and tissue-specific patterns • Correlations among chromatin state, gene expression and function Construct cell- and tissue-specific cis-regulatory maps • Mapping chromatin regulators and TFs that control regulatory elements • Integrating tissue-specific expression with chromatin state and TF data Histone modification signal profiles Transcription factors Pol II Histone mods From Park (2009) Nat Rev Genet 10:669 Quantitative analysis of ChIP-seq signal profiles HeLa K562 Histone modification signal Signal at 20,000 bound sites HeLa Sites strongly marked in HeLa Sites strongly marked in both Clustering Sites strongly marked in K562 Identifying chromatin state differences among cell types K-means clustering of histone modification signals at promoters across cell types • Given a predetermined number of clusters (k), each promoter is assigned to cluster with the nearest mean Heintzman et al. Nature 459:108 (2009) • Chromatin signatures at promoters tend to be more common across tissues • Enhancers tend to be more tissue-specific Identifying chromatin state differences among cell types Enhancers Heintzman et al. Nature 459:108 (2009) Cell-type specific enhancer activation correlates with cell-type specific gene expression Heintzman et al. Nature 459:108 (2009) Integrated analysis of histone modifications identifies functional ‘states’ Promoter Chromatin mark observation frequency Enhancer ChromHMM: infer chromatin states from combinations of histone marks • ‘emission’ probabilities associated with each chromatin state • ‘transition’ probabilities – frequency in which chromatin states occur in spatial relationships with each other along genome Ernst et al., Nature 473:43 (2011) Mapping and analysis of chromatin state dynamics in nine human cell types Cell types: Marks: • H1 ESC • H3K4me3 (promoter/enhancer) • K562 (erythrocyte derived) • H3K4me2 (promoter/enhancer) • GM12878 (B-lymphoblastoid) • H3K4me1 (enhancer) • HepG2 (hepatocellular carcinoma) • H3K9ac (promoter/enhancer) • HUVEC (umbilical vein endothelium) • H3K27ac (promoter/enhancer) • HSMM (skeletal muscle myoblasts) • H3K36me3 (transcribed regions) • NHLF (lung fibroblast) • H4K20me1 (transcribed regions) • NHEK (epidermal keratinocytes) • H3K27me3 (Polycomb repression) • HMEC (mammary epithelium) • CTCF Ernst et al., Nature 473:43 (2011) Mapping and analysis of chromatin state dynamics in nine human cell types Ernst et al., Nature 473:43 (2011) Chromatin states vary across cell types Chromatin states at WLS: Ernst et al., Nature 473:43 (2011) Genetic drivers of gene regulatory variation Furey and Sethupathy, Science Disease-associated SNPs disrupt putative cell-type specific enhancers Ernst et al., Nature 473:43 (2011) Chromatin state segmentation maps of the human genome Summary • Histone modifications reveal where functions are encoded in the genome • Combinatorial analysis of histone modifications across biological states identifies tissue-specific chromatin states • Tissue-specific chromatin states reveal tissue-specific enhancers, promoters, etc. • Integrated analyses of chromatin states provide functional annotations for predicting the effects of genetic variation