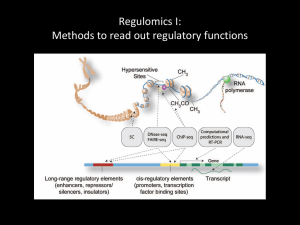
Task 1. Histone modification ChIP-seq pre
advertisement

Task 1. Histone modification ChIP-seq pre-processing (months 1 – 6) Dr. Dean Tang’s lab at MD Anderson Cancer Center (MDACC) will independently perform ChIP-seq experiments for the 5 proposed histone modifications in LNCaP system and LAPCS9 xenograft. Please note that the work at DR. Tang’s lab at MDACC is covered under separate funding thus is not part of this project. We will examine the quality of individual ChIP-seq experiment and further implement the proposed data processing pipelinedetect differential histone modifications between PCSCs and nnonPCSCs. We are aware that variability within a group of clinical samples can be huge, so we will increase sample size when necessary to obtain the statistically significant results. 1a. Performing ChIP-seq quality control analyses, pre-processing and visualization (months 1 - 6) 1b. Implementing the signal extraction data pipeline (months 1 - 6) Task 2: Identification of combinatorial histone modification patterns for PCSCs (months 6 – 36) 2a. Identifying significantly differentiated signals between PCSCs and non-PCSCs for each histone modification mark. (months 6 - 12) 2b. More generally, integrating different histone modification marks and identifying combinatorial histone modification patterns in PCSCs. (Months 12 -36) Task 3: Identification of H3K4me3 super promoter patterns in PCSCs and nonPCSCs (months 12 – 24) 3a. Identifying significantly differentiated signals between PCSCs and non-PCSCs for each histone modification mark. (months 6 - 12) 3b. Implementing the peak combining algorithm to detect elongated H3K4me3 peak patterns. (Months 12 -24) Task 4: Explore biological/clinical functions for gene sets with PCSCs specific epigenetic signatures (months 18 – 36) We will perform functional gene set analyses to discover potential pathways, gene ontology (GO) terms and potential regulatory mechanisms relevant to CRPC. We will perform correlation analyses with public prostate cancer data, including expression, DNA methylation levels, sequence features, etc. We will provide a subset of key aberrations that are useful in differentiating between PCSCs and non-PCSCs, and follow up with our collaborators for experimental validations. We will achieve four milestones: 1) We will design, test, and validate an advanced bioinformatics pipeline for the identification of super promoter H3K4me3 peak patterns using ChIP-seq (month 18), by developing novel methodologies as well as integrating existing approaches where appropriate. 2) We will design, test, and validate an advanced bioinformatics pipeline for the identification of cancer specific alternative splicing using (month 18), by developing novel methodologies as well as integrating existing approaches where appropriate. 3) We will design, test, and validate a PCSC/non-PCSC classification framework based on integrated genome-wide data (month 30). 4) We will provide a subset of key aberrations that are useful in differentiating between PCSCs and non-PCSCs (month 36).