Here
advertisement
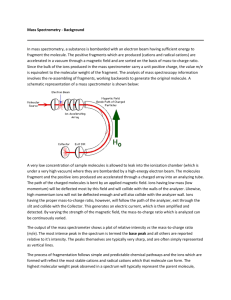
Theodore Alexandrov, Michael Becker, Sören Deininger, Günther Ernst, Liane Wehder, Markus Grasmair, Ferdinand von Eggeling, Herbert Thiele, and Peter Maass Background on MS Imaging and goals of paper Methods Results Conclusions and Criticism Background on MS Imaging and goals of paper Methods Results Conclusions and Criticism In the words of All-Mighty Wikipedia: Mass spectrometry imaging is a technique used in mass spectrometry to visualize the spatial distribution of e.g. compounds, biomarker, metabolites, peptides or proteins by their molecular masses. Or in images: To propose a new procedure for spatial segmentation of MALDI-imaging datasets. This procedure clusters all spectra into different groups based on their similarity. This partition is represented by a segmentation map, which helps to understand the spatial structure of the sample. (it is MS Imaging after all) Current multivariate algorithm (PCA) are not meant for MS data and cannot be used to directly interpret the data. Current clustering algorithm do not take in account spatial information. Here, we assume that spectra close to each other should be similar. Background on MS Imaging and goals of paper Methods Results Conclusions and Criticism Rat brain coronal section ◦ ◦ ◦ ◦ 80 µm raster 200 laser shots per position; 20185 spectra Data acquired: 2.5 kDa-25 kDa Data considered: 2.5 kDa-10 kDa; 3045 points Section of neuroendocrine tumor (NET) invading the small intestine ◦ ◦ ◦ ◦ 50 µm raster 300 laser shots per position; 27360 spectra Data acquired:1 kDa-30 kDa Data considered: 3.2 kDa-18kDa; 5027 points Baseline correction ◦ TopHat algorithm, minimal baseline width set to 10%, default in ClinProTools No normalization No binning ASCII -> Matlab Part1: conventional peak picking applied to each 10th spectrum. Select 10 peaks. ◦ Orthogonal Matching Pursuit (OMP) because it is fast and simple ◦ Gaussian kernel deconvolution Part 2: keep consensus peaks: ◦ Only keep peaks that appear in at least 1% of the considered spectra ◦ Omit spurious peaks Imaging dataset is a reduced datacube with 3 coordinates: x, y, m/z (reduced in m/z dimension by peak picking) MALDI-imaging data is noisy Must be able to keep fine anatomical or histological details Grasmair modification of Total Variation minimizing Chambolle algorithm ◦ Parameter θ between 0.5 and 1: smoothness of resulting image Total variation (TV) ~ sum of absolute differences between neighboring pixels Chambolle algorithm searches for an approximation of the image with small TV Chambolle algorithm => smoothness adjusted globally by manually choosing a parameter Grasmair locally adapts denoising parameter of Chambolle Specify number of cluster a-priori High Dimensional Discriminant Clustering (HDDC) ◦ Available in Matlab tool box ◦ Each cluster is modeled by a Gaussian distribution of its own covariance structure. ◦ HDDC developed for high-dimensional data (d > 10) ◦ Note: In Matlab HDDC = high-dimensional data clustering Background on MS Imaging and goals of paper Methods Results Conclusions and Criticism used 2019 spectra out of 20185 (10%) potential peaks: 373 peaks (red triangles) consensus peaks: 110 peaks (green triangles) ◦ Present in at least 20 spectra out of the 2019 (1%) Discarded peaks mostly in low m/z regions Hypothesize they are noise peaks because MALDI imaging spectra have high baseline in low m/z region. OMP successfully detects major peaks Gaussian function provides reasonable approxima tion of peak shape Strong noise Noise variance changes within m/z image and between m/z images Noise variance is linearly proportional to peak intensity Apply Grasmair method to selected 110 consensus peaks Efficiently removes the noise while not smoothing out edges Shows anatomical features Restricted to spatial resolution of MALDIimaging dataset No denoising: borders do not match as well 3x3 median smoothing: bad edge preservation 5x5 median smoothing: lose many regions Find mass values expressed in region 3 main parameters in addition to peak width ◦ Portion of spectra considered for peak picking (each 10th spectrum) ◦ Number of peaks selected for each spectrum (10 peaks) ◦ Percentage of spectra where peak is found for consensus peak list (1%) Robust to changes of second and third parameter 5 0.1% 1% 5% 10 20 peaks Increase of parameter 1 can be compensated by higher value for parameter 2 Each 20th spectrum Each 5th spectrum Segmentation maps for ◦ 3 levels of denoising (0.6, 0.7, 0.8) ◦ 3 number of clusters (6, 8, 10) Decrease in number of clusters merge features Too much denoising causes loss of structure details Background on MS Imaging and goals of paper Methods Results Conclusions and Criticism Peak picking: usually done on mean spectrum ◦ 1% consensus better for peaks in small spatial area Edge-preserving denoising ◦ One study with average moving window and one study posthoc to improve classification Clustering methods ◦ HDDC better results than k-means but significantly slower ◦ Currently, mostly hierarchical clustering = memory intensive Importance to cancer studies ◦ Represents a proteomic functional topographic map Didn’t explain why they got rid of part of the range for which the data was acquired Dataset reduction by peak picking ◦ done initially on per spectrum basis, it may get rid of lower abundance peaks which still show interesting image ◦ Also, because the peak must be present in 1% of the 10% selected spectra, can miss smaller regions of interest if bad selection of 10% Highly parameterized + slow running time would make it hard to run many trials