Affinty chromatography
advertisement

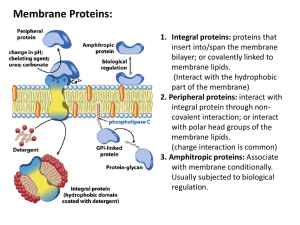
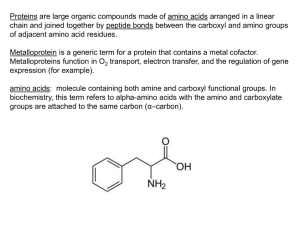
In the Name of GOD Affinity Chromatography Introduction A goal of biochemistry is to separate and identify chemical compounds. chromatography is one of the most effective techniques for accomplishing this. In chromatography, substances are placed in a system consisting of two physically distinguishable components -a mobile phase and a stationary phase-and molecular species separate because they differ (many of them only slightly) in their distribution between these two phase. There are many kinds of chromatography: Adsorption Partition Ion-Exchange Molecular Sieve Affinity Column, paper, thin-layer and gas chromatography. 2 Historical Perspective of Affinity Chromatography * The German pharmacologist Emil Starkenstein in paper published in 1910 on the influence of chloride on the enzymatic activity of liver-amylase was generally considered to be responsible for the first experimental demonstration of the biospecific adsorption of an enzyme onto a solid substrate, in this case, starch. * Not long after, Willstatter et al. appreciably enriched lipase by selective adsorption onto powdered stearic acid. * Affinity chromatography as it is known today was introduced in 1968 by Cuatrecasas et al. 4 Affinity Chromatography The technique offers high selectivity, hence high resolution, and usually high capacity for the proteins of interest. Purification that would otherwise be time-consuming, difficult or even impossible using other techniques can often be easily achieved with affinity chromatography. The technique can be used to separate active biomolecules from denatured or functionally different forms, to isolate pure substances present at low concentration in large volumes of crude sample and also to remove specific contaminants. 5 6 7 Affinity Chromatography Affinity chromatography separates proteins on the basis of a reversible interaction between a protein and a specific ligand coupled to a chromatography matrix. The kinds of Elution pH Elution Ionic Strength Elution Reduced Polarity of Eluent Competitive Elution Chemotropic Eluents 8 Elution 9 10 Purification steps 13 14 Binding: buffer conditions are optimized to ensure that the target molecules interact effectively with the ligand and are retained by the affinity medium as all other molecules wash through the column. Elution: buffer conditions are changed to reverse (weaken) the interaction between the target molecules and the ligand so that the target molecules can be eluted from the column. Wash: buffer conditions that wash unbound substances from the column without eluting the target molecules or that re-equilibrate the column back to the starting conditions (in most cases the binding buffer is used as a wash buffer). Ligand coupling: covalent attachment of a ligand to a suitable pre-activated matrix to create an affinity medium. Pre-activated matrices: matrices which have been chemically modified to facilitate the coupling of specific types of ligand. 15 Affinity Medium Matrix The Kinds of matrix Ligand Ligamd Immobilization Spacer Arm 16 Matrix Proper selection of a matrix or carrier for the ligands is of decisive importance for the successful application of stereospecific adsorption. 1. 2. 3. 4. 5. 6. 7. 8. 9. Insolubility Sufficient permeability High rigidity and suitable particle form Zero adsorption capacity Chemical stability under the conditions required for adsorption, desorption and regeneration Chemical reactivity allowing ligands and spacers to be introduced Resistance toward microbial and enzymatic attack Good flow properties for rapid separation An open pore structure ensures high capacity binding even for large biomolecules. 17 No matrix support is ideal in all these respects. Porous glass Cellulose Polyacrylamide Agarose 18 Ligand The selection of the ligand for affinity chromatography is influenced by two factors: o o the ligand must exhibit specific and reversible binding affinity for the target substance and it must have chemically modifiable groups that allow it to be attached to the matrix without destroying binding activity. The dissociation constant (kD) for the ligand-target complex should ideally be in the range 10-4 to 10-8 M. 19 Ligand Immobilization Sience 1970, a large number of methods have been developed for coupling ligands to matrix materials. The most common procedure is to link a coupling agent to the matrix material and then add the ligand. It is important to mention that, after coupling of the desired ligand, reactive Y Activation Step: groups may still be present. Deactivation may occur by spontaneous hydrolysis but, if this is not the case, coupling with a low molecular weight substance. Glycine, neutral dipeptides, and ethanolamine are deactivating substances that should be considered. Coupling Step: 20 Methods for Immobilization 1. Cyanogen Bromide Coupling 2. Bisoxirane Coupling 3. Divinylsulfone Coupling 21 1. Cyanogen Bromide Coupling 1,2-Diols are especially liable to react with cyanogen halides to form cyclic imino carbonates. In the coupling step a substance containing amino groups will form at least three products. Activation Step: Coupling Step: 22 This reaction is extremely useful in coupling enzymes, coenzymes, inhibitors, antigen, antibodies, nucleic acids and most proteins to agarose. Although most applications of cyanogen bromide coupling have involved agarose and cross-linked agarose, other hydroxylcontaining polymers may also be converted to biospecific adsorbents by coupling of suitable ligands in the same manner. 23 2. Bisoxirane Coupling Bisoxiranes (bisepoxides) are particularly useful reagents for introducing low molecular weight ligands through amino or hydroxyl groups. 24 3. Divinylsulfone Coupling The vinyl groups introduced into the matrix are more reactive than are the oxirane groups. They will thus couple to amines, alcohols, and phenols at lower temperatures and at lower pH than the oxirane. 25 Spacer Arm The binding site of a target protein is often located deep within the molecule and an affinity medium prepared by coupling small ligands, directly to matrix may exhibit low binding capacity due to steric interference i.e. the ligand is unable to access the binding site of the target molecule. * The length of the spacer arm is critical. * when using small ligands (Mr < 5 000) there is a risk of steric hindrance between the ligand and the matrix that restricts the binding of target molecules. In this case, select a pre-activated matrix with a spacer arm. For ligands with Mr > 5 000 no spacer arm is necessary. 26 Ligand Design 1. Protein-Structure-Based Design 2. Protein-Function-Based Design 27 Ligand Disign The rapid growth of bioinformatics and molecular docking and the introductionDesign of combinatorial methods for 2. 1.techniques Protein-Function-Based Protein-Structure-Based Design systematic generation and screening of large numbers of novel The strategy for the rational design affinity ligands involves This approachhas is applied where the the threeof dimensional ofof the compounds, made feasible rapid and efficient structure generation target is notchromatography. available and is relies thetarget incorporation retrieving information aboutonthe protein of from ligandsprotein forstructural affinity certain structural features on the ligand. suitable databases and identifying a potential binding site on the 1. 2. 3. A certain required molecular shape. protein. Specific functional For example, forgroup. the design of ligands for A structural model derived from the combination of structural moieties which •L-Lactate Dehydrogenase are known substrates, inhibitors, effectors or cofactors. •Glutathione S-transferase •Galactose Oxidase •Galactose Dehydrogenase •Elastase. 28 29 30 31 32 33 Biomimetic Dyes 34 Biomimetic Dyes as Affinity Chromatography In 1970, immobilized triazine dyes, particularly Cibacron blue, have been used as affinity chromatography tools for protein and enzyme purification. The low cost of these dyes, their ease of immobilization and resistance to biological and chemical degradation, and the high protein-binding capacity of the corresponding adsorbents. Although textile dyes, in some case, interact with proteins with remarkable degrees of specificity, their interaction with a large number of seemingly unrelated proteins inevitably compromises their protein binding specificity and endow these molecules with a serious drawback. 35 Another strategy which canofcope with the of textile One way to cope with lack specificity ofdrawback immobilized dyes isdyes, to is tospecific design eluents new dye-ligands of improved affinity andthe specificity use which allow to elute specifically target for the target protein. protein with minimal contamination. In principle this beknown achieved by designing synthetic This approach is can better as affinity elution. An dyes which mimic the structure and binding of chromatography, natural biologicalutilizing ligands of alternative method of affinity thethe targetedofprotein. formation specific complex of the macromolecule under isolation with an affinity ligand, is biospecific elution from non-specific adsorbents and in some instances as “specific elution by the substrate”. 36 First Generation Biomimetic Dyes Early in the 1980s, the time the first biomimetic dye was under design, development and assessment, were not available advanced molecular modeling software packages for application on a personal workstation, Therefore, biomimetic dye design was relying on the known binding preference of the target enzyme for natural ligands, X-ray crystallography data, and other available useful biochemical information. o Alkaline phosphatase o Alcohol dehydrogenase o Trypsin 37 Second Generation Biomimetic Dyes Design by computer aided molecular modeling and use of bioinformatics. In fact this computational technology, an area of bioinformatics, has marked a new era in ligand design. L-malate dehydrogenase L-lactate dehydrogenase 38