Prof. Kamakaka`s Lecture 4 Notes
advertisement

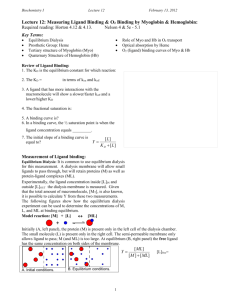
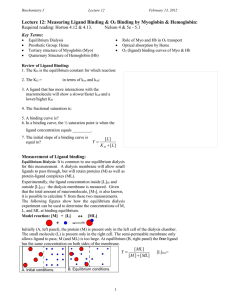
Ligands and reversible binding Ligands Kinetic experiments study the rate at which reactions happen.- how conc of reactant and product change as funct of time. Rate of reaction is slope. Rate of reaction decreases as reaction proceeds. Chemical kinetics Equilibrium Equilibrium experiments study how conc of reaction products change as function of reactant concentrations. A+B<---->AB. Increasing amount of A is titrated against fixed amount of B and equilibrium conc of product AB determined. Thermodynamics Rate constants and equilibrium constant • Consider a process in which a ligand (L) binds reversibly to a site in the protein (P) ka kd ka [PL] = Ka= kd [P][L] In practice, we can often determine the fraction of occupied binding sites Q= [PL] [PL]+[P] [PL] Ka= [P][L] Q= Bound protein Total protein [P][L] Kd= [PL] [L] [L]+ Kd • The fraction of bound sites depends on the free ligand concentration and Kd • In a typical experiment, ligand concentration is the known independent variable Ligand binding to protein The fraction of ligand-binding sites occupied Q plotted against conc of free ligand • Interaction strength can be expressed as: – association (binding) constant Ka, units M‐1 – dissociation constant Kd, units M, Kd = 1/Ka – interaction (binding) free energy DGo, units: kJ/mol Definitions: – DGo = DHo ‐TDSo : enthalpy and entropy – Ka = [PL]/[P][L] - Kd=[P][L]/[PL] • Relationships: – DGo = ‐RT ln Ka = RT ln Kd (RT at 25 oC is 2.48 kJ/mol) • Magnitudes – Strong binding: Kd < 10 nM – Weak binding: Kd > 10 uM Myoglobin Myoglobin Heme Myoglobin, Histidine and oxygen Carbon Monoxide • CO has similar size and shape to O2; it can fit to the same binding site • CO binds over 20,000 times better than O2 because the carbon in CO has a filled lone electron pair that can be donated to vacant d-orbitals on the Fe2+ • Myoglobin Protein pocket decreases affinity for CO, but it still binds about 250 times better than oxygen • CO is highly toxic as it competes with oxygen. It blocks the function of myoglobin, hemoglobin, and mitochondrial cytochromes that are involved in oxidative phosphorylation Myoglobin Oxygen binding Hemoglobin Oxygen binding. Hemoglobin cycle XXX Myoglobin and Hemoglobin structures T and R structures T and R states Hi K CO2 CO2 CO2 CO2 b a K Hi T state Tissues = low pH Low O2 conc (4 kPa) No oxygen bound H+/CO2 bound a O O O O b K Hi b Hi K a b a R state Lungs = high pH High O2 (13.3 kPa) Oxygen bound No H+/CO2 bound Allosteric Interactions Heterotropic Protein Protein Ligand Modulator binds Ligand induces change in protein and binds protein Modulator induces change Homotropic Ligand binds protein Myoglobin and Hemoglobin O2 binding % bound by Ligand 100 50 Myo Hb Ligand concentration Slope and degree of co-operativity Slope is the measure of the degree of co-operativity For Hb in the low and high affinity states, their slopes indicate no-cooperativity For Hb in the intermediate states, the slope indicates high cooperativity Co-operativity Molecular models for cooperativity XXXXXXX Induced fit