Essential amino acids
advertisement

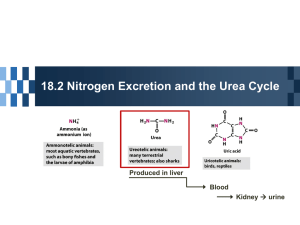
蛋白质的分解代谢 Protein Degradation and Amino Acids Metablism Contents Protein degradation Amino Acid Degradation Biosynthesis of amino acids I. Protein Degradation Biological Functions of Proteins Enzymes Transport proteins Nutrient and storage proteins Contractile or motile proteins Structural proteins Defense proteins Regulatory proteins Other proteins Nitrogen balance • Zero or total nitrogen balance: the intake = the excretion (adult) • Positive nitrogen balance: the intake > the excretion (during pregnancy, infancy, childhood and recovery from severe illness or surgery ) • Negative nitrogen balance: the intake < the excretion (following severe trauma, surgery or infections. Prolonged periods of negative balance are dangerous and fatal. ) Classification of amino acids • non-essential amino acids - can be synthesized by an organism - usually are prepared from precursors in 1-2 steps • Essential amino acids *** - can not be made endogenously - must be supplied in diet Nonessential Essential Alanine Arginine* Asparagine Histidine * Aspartate Valine Cysteine Lysine Glutamate Isoleucine Glutamine Leucine Glycine Phenylalanine Proline Methionine Serine Threonine Tyrosine Tyrptophan *The amino acids Arg, His are considered “conditionally essential” for reasons not directly related to lack of synthesis and they are essential for growth only Degradation of dietary proteins Degradation of proteins 1. Degraded by ubiquitin(泛素) label 2. Degraded by the protease and the peptidase in the Lysosome(溶酶体) 1. Degraded by ubiquitin(泛素) label Ubiquitin, a extremely well conserved 76residue protein, Ubiquitin binds lysine side chain Degrade abnormal protein of her own Targets for hydrolysis by proteosomes in cytosol and nucleus ATP required 2. Degraded by the protease and the peptidase in the Lysosome(溶酶体) non- ATP required the hydrolysis-selective are bad Degrade adventive protein The ubiquitin degradation pathway ATP AMP+PPi E1-S(ubiquitin) E3 E2-SH E2-SE1-SH E1-SH E2-SH E1:activiting enzyme E2:carrier protein E3:ligase ubiquitinational protein 19S regulate substrate ATP ATP 20S Proteasome 26S Proteasome II. Amino acids Degradation The catabolism of amino acids I. Deamination A. Transamination B. Oxidative deamination C. Combined Deamination A. Transamination Transamination by Aminotransferase (transaminase) always involve PLP coenzyme (pyridoxal phosphate) reaction goes via a Schiff’s base intermediate all transaminase reactions are reversible Transamination aminotransferases B. Oxidative Deamination • L-glutamate dehydrogenase (in mitochondria) C. Combined Deamination 1. Transamination + Oxidative Deamination ? NH3 AA Asp IMP -Keto glutarate H2O aminotransferasesAST -Keto acid 2. Transamination + purine nucleotide cycle Oxaloacetate malate fumarate AMP II. Decarboxylation The decarboxylation of AAs produce some neurotransmitters’ precursors – bioactive amines -aminobutyric acid (GABA) Glutamine can be decarboxylated in a similar PLP-dependent fashion, outputting -aminobutyric acid (neurotransmitter, GABA) COOH COOH (CH2)2 CHNH2 COOH L-Glu L-Glu decarboxylase – CO2 (CH2)2 CH2NH2 GABA H N CH 2 C COOH NH NH2 L-Histidine Histamine 强烈的血管舒张剂。增加 CH 2CH 2NH 2 血管的通透性,降低血压, 甚至死亡。 N NH Histamine III. The metabolism of α-ketoacid Biosynthesis of nonessential amino acids TCA cycle member + amino acid α-keto acid + nonessential amino acid A source of energy (10%) ( CO2+H2O ) Glucogenesis and ketogenesis Fate of the C-Skeleton of Amino Acids Ⅳ . ammonia metabolism Fix ammonia onto glutamate to form glutamine and use as a transport mechanism Transport ammonia by alanine-glucose cycle and Gln regeneration Excrete nitrogenous waste through urea cycle Transportation of ammonia • alaninie - glucose cycle * • regenerate Gln Alanine-Glucose cycle In the liver alanine transaminase tranfers the ammonia to α-KG and regenerates pyruvate. The pyruvate can then be diverted into gluconeogenesis. This process is refered to as the glucosealanine cycle. Gln regeneration Urea synthesis Synthesis in liver (Mitochondria and cytosol) Excretion via kidney To convert ammonia to urea for final excretion The urea cycle: CO2 + NH3 + H2O 2ATP N-乙酰谷氨酸 2ADP+Pi 线粒体 氨基甲酰磷酸 Pi 瓜氨酸 鸟氨酸 瓜氨酸 ATP AMP + PPi 天冬氨酸 鸟氨酸 尿素 胞液 精氨酸代 琥珀酸 草酰乙酸 精氨酸 延胡索酸 苹果酸 α-酮戊 二酸 谷氨酸 氨基酸 α-酮酸 UREA CYCLE (liver) 1. Overall Reaction: NH3 + HCO3– + aspartate + 3 ATP + H2O urea + fumarate + 2 ADP + 2 Pi + AMP + ppi 2. Requires 5 enzymes: 2 from mitochondria and 3 from cytosol Regulation of urea cycle The intake of the protein in food:the intake↑↑urea synthesis AGA:CPS I is an allosteric enzyme sensitive to activation by N-acetylglutamate(AGA) which is derived from glutamate and acetyl-CoA. All intermediate products accelerate the reaction Rate-limiting enzyme of urea cycle is argininosuccinate synthetase(精氨酸代琥珀酸合成酶) The Urea Cycle is Linked to the Citric Acid Cycle NH4+ III. Biosynthesis of Amino acids Ammonium Ion Is Assimilated into Amino Acids Through Glutamate and Glutamine Major Ammonium ion carrier Biosynthesis of Amino Acids