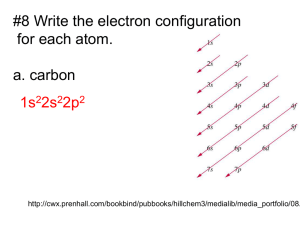
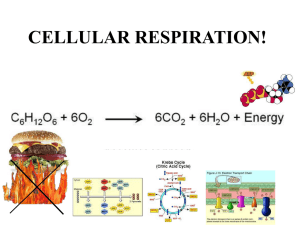
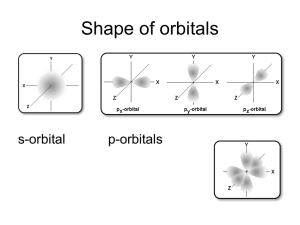
Oxidation
advertisement

Chapter 19 Oxidative Phosphorylation and Photophosphorylation Oxidative Phosphorylation In mitochondria Reduction of O2 to H2O with electrons from NADH or FADH2 Independent on the light energy Photophosphorylation In chloroplast Oxidation of H2O to O2 with NADP+ as electron acceptor Dependent on the light energy Oxidative Phosphorylation vs. Photophosphorylation Similarities Flow of electrons through a chain of membrane-bound carriers (Downhill: exogernic process) Proton transport across a proton-impermeable membrane (Uphill: endogernic process) Free energy from electron flow is coupled to generation of proton gradient across membrane Transmembrane electrochemical potential (conserving free energy of fuel oxidation) “Chemiosmotic theory by Peter Mitchell (1961)” Proton gradient as a reservoir of energy generated by biological oxidation ATP synthase couples proton flow to ATP synthesis Oxidative Phosphorylation 19.1 Electron-Transfer Reactions in Mitochondria Mitochondria Site of oxidative phosphorylation Eugene Kennedy and Albert Lehninger (1948) Structure Outer membrane Free diffusion of small molecules (Mr < 5,000) and ions through porin channels Inner membrane Impermeable to most small molecules and ions (protons) Selective transport Components of the respiratory chain and the ATP synthase Mitochondria matrix Contain enzymes for metabolism Pyruvate dehydrogenase complex Citric acid cycle b-oxidation Amino acid oxidation Electron transfer in biological system Types of electron transfer in biological system Direct electron transfer : Fe3+ Fe2+ Hydrogen atom (H+ + e-) Hydride ion (:H-) Organic reductants * Reducing equivalent A single electron equivalent transferred in an redox reaction Types of electron carriers NAD(P)+ FAD or FMN Ubiquinone (coenzyme Q , Q) Cytochrome Iron-sulfur proteins NAD(P)+ & FAD/FMN ; universal electron acceptors NAD(P)+ -Cofactors of dehydrogenases (generally) -Electron transfer as a form of :H-Low [NADH]/[NAD+] catabolic reactions -High [NADPH]/[NADP+] anabolic reactions -No transfer into mito matrix -Shuttle systems (inner mito membrane) FAD/FMN (flavin nucleotides) -Tightly bound in flavoprotein (generally) -One (semiquinone) or two (FADH2 or FMNH2) Partial reduction; 450nm absorption Full reduction; 360nm absorption electron accept -High reduction potential (induced by binding to protein) Full oxidation; 370 & 440 nm absorption Membrane-bound electron carriers ; Ubiquinone Coenzyme Q or Q Lipid-soluble benzoquinone with long isoprenoid side chain Accept one (semiquinone radical; •QH) or two electrons (ubiquinol; QH2) Freely diffusible within inner mito membrane Shuttling reducing equivalents between less mobile electron carriers Coupling electron flow to proton movement Membrane-bound electron carriers ; Cytochromes Iron-containing heme prosthetic group 3 classes of Cyt in mitochondria (depending on differences in light-absorption spectra) ; a (near 600nm), b (near 560nm), c (near 550nm) Cyt c - Covalently-attached heme through Cys - Soluble protein associated with outer surface of inner mito membrane Membrane-bound electron carriers ; Iron-sulfur proteins Irons associated with inorganic S or S of Cys One electron transfer by redox reaction of one iron atom > 8 Fe-S proteins involved in mito electron transfer Reduction potential of the protein : -0.65 V ~ +0.45 V Determining the Sequence of Electron Transfer Chain Based on the order of standard reduction potential (E’°) Electron flow from lower E’° to higher E’° NADH Q Cyt b Cyt c1 Cyt c Cyt a Cyt a3 O2 Determining the Sequence of Electron Transfer Chain Reduction of the entire chain of carriers sudden addition of O2 Spectroscopic measurement of oxidation of each electron carriers Closer to O2 faster oxidation Inhibitors Blocking the flow of electrons Before/after the inhibited step : fully reducted/ fully oxdized Electron Carriers in multienzyme complex Membrane-embedded supramolecular complexes (organized in mito respiratory chain) Complex I : NADH Q Complex II : Succinate Q Complex III : Q Cyt c Complex IV : Cyt to O2 Separation of functional complexes of respiratory chain Electron Carriers in multienzyme complex Path of electrons from various donors to ubiquinone Complex I : NADH:ubiquinone oxidoreductase (NADH dehydrogenase) 42 polypeptide chains FMN-containing flavoprotein > 6 iron sulfur centers Functions : proton pump driven by the energy from electron transfer Exergonic transfer of :H- from NADH and a proton from the matrix to Q NADH + H+ + Q NAD+ + QH2 Endergonic transfer 4 H+ from the matrix to the intermembrane space NADH + 5HN+ + Q NAD+ + QH2 + 4Hp+ Inhibitors : e- flow from Fe-S center Amytal (a barbiturate drug) Rotenone (plant, insecticide) Piericidin A (antibiotic) Complex II : Succinate Dehydrogenase Only membrane-bound enzyme in the citric acid cycle Structure 4 subunits C and D : transmembrane side Heme b : preventing electron leakage to form reactive oxygen species Q binding site A and B : matrix side Three 2Fe-2S centers FAD Binding site of succinate Electron passage : entirely 40 Å long (< 11 Å of each step) Electron transfer from Glycerol 3phosphate & fatty acyl-CoA Electron from fatty acyl-CoA FAD electron-transferring flavoprotein (ETF) ETF: ubiquinone oxidoreductase Q Electron from glycerol 3-phosphate FAD in glycerol 3-phosphate dehydrogenase Q Shuttling reducing equivalents from cytosolic NADH into mito matrix ; glycerol 3-phosphate dehydrogenase Complex III: Cyt bc1 complex (Q:Cyt c oxidoreductase) e- transfer (ubiquinol (QH2) Cyt c) H+ transfer (matrix intermembrane space) Dimer of identical monomers (each with 11 different subunits) Functional core of each monomer; cyt b (2 heme; bH & bL) + Rieske iron-sulfur protein (2Fe-2S center) + cyt c1 (heme c1) Complex III: Cyt bc1 complex (Q:Cyt c oxidoreductase) Two binding sites for ubiquinone ; Q N & QP Antimycin A: binding at QN block e- flow (heme bH Q) Myothiazol: binding at QP block e- flow (QH2 Rieske iron-sulfur protein) Cavern (space at the interface between monomers) ; QN & QP are located Q cycle in complex III Two stages 1st stage; Q (on N side) semiquinone radical 2nd stage; semiquinone radical QH2 Complex IV : Cytochrome Oxidase e- transfer from cyt c to O2 H2O Structure; 13 subunits Subunit II; 2 Cu ions complexed with –SH of 2 Cys (CuA) 1st binuclear center Subunit I; 2 heme groups, a & a3 Cu ion (CuB) a3 + CuB 2nd binuclear center Complex IV : Cytochrome Oxidase Electron transfer Cyt c CuA heme a heme a3-CuB center O2 4 Cyt c (red) + 8 HN+ + O2 4 cyt c (ox) + 4Hp+ + 2 H2O 4HN+ as substrate, 4HN+ for pumping out