PowerPoint-presentation
advertisement
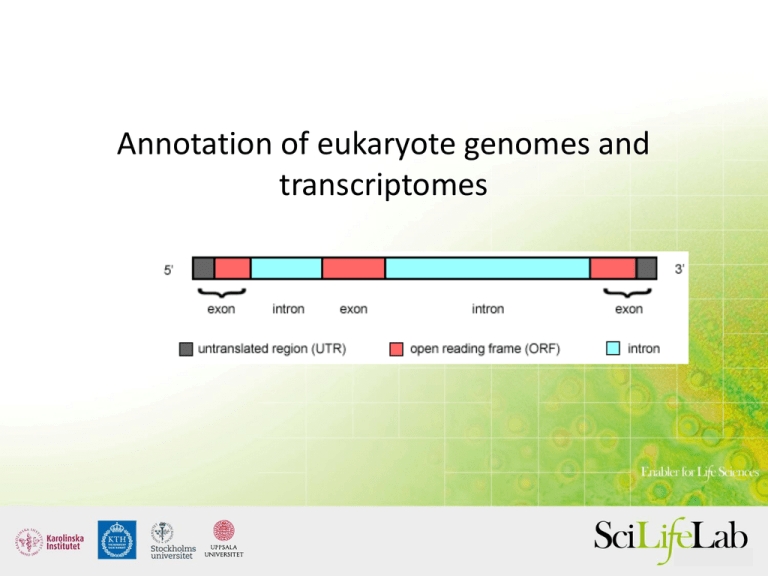
Annotation of eukaryote genomes and transcriptomes Some possibilities and some pitfalls What is annotation? • Identification of regions of interest in sequence data From a genome… …to an annotated gene An annotation GTF file A reminder, you know this already… There are two major parts of annotation • 1) Structural: Find out where the regions of interest (usually genes) are in the genome and what they look like. How many exons/introns? UTRs? Isoforms? • 2) Functional: Find out what the regions do. What do they code for? The challenge: Which one is correct? Transcriptomes are different but have their own challenges • No introns, but where are the start and stop codons? • Still needs functional annotation Why is annotation important? RNA-seq reads Genome Data used - Proteins • Conserved in sequence => conserved annotation with little noise • Proteins from model organisms often used => bias? • Proteins can be incomplete => problems as many annotation procedures are heavily dependent on protein alignments >ENSTGUP00000017616 pep:novel chromosome:taeGut3.2.4:8_random:2849599:2959678:-1 gene:ENSTGUG00000017338 transcript:ENSTGUT000000180 RSPNATEYNWHHLRYPKIPERLNPPAAAGPALSTAEGWMLPWGNGQHPLLARAPGKGRER DGKELIKKPKTFKFTFLKKKKKKKKKTFK >ENSTGUP00000017615 pep:novel chromosome:taeGut3.2.4:23_random:205321:209117:1 gene:ENSTGUG00000017337 transcript:ENSTGUT00000018017 PDLRELVLMFEHLHRVRNGGFRNSEVKKWPDRSPPPYHSFTPAQKSFSLAGCSGESTKMG IKERMRLSSSQRQGSRGRQQHLGPPLHRSPSPEDVAEATSPTKVQKSWSFNDRTRFRASL RLKPRIPAEGDCPPEDSGEERSSPCDLTFEDIMPAVKTLIRAVRILKFLVAKRKFKETLR PYDVKDVIEQYSAGHLDMLGRIKSLQTRVEQIVGRDRALPADKKVREKGEKPALEAELVD ELSMMGRVVKVERQVQSIEHKLDLLLGLYSRCLRKGSANSLVLAAVRVPPGEPDVTSDYQ SPVEHEDISTSAQSLSISRLASTNMD Data used - rna-seq • Should always be included in a annotation project • From the same organism as the genomic data => unbiased • Can be very noisy (tissue/species dependent), can include pre-mRNA • PASA, or some other filtering method, often needed Adding RNAseq - Spliced reads Pre-mRNA Includes everything that is transcribed Stranded rna-seq Bring home message: Combine data General recommendations • Always combine different types of evidence! • One single method is not enough! • Use Maker! Types of evidence 1. De novo gene finders, some in combination with RNA-seq 2. Transcripts assembled from mapped RNAseq reads 3. Assemble transcripts de novo followed by mapping 4. Lifting over annotations from a closely related genome 5. Protein alignments De novo gene prediction • Commonly used programs: Genemark-ES, Augustus, Glimmer-HMM, Snap, Genscan, FGENESH… • Uses HMM-models to figure out how introns, exons, UTRs etc. are structured • These HMM-models need to be trained! Different training files De novo gene prediction • Commonly used programs: Genemark-ES, Glimmer-HMM, Snap, Genscan, FGENESH… • Uses HMM-models to figure out how introns, exons, UTRs etc. are structured • These HMM-models need to be trained! • glimmerhmm_linux fasta.file -d GlimmerHMM/trained_dir/arabidopsis/ • Each program comes with information on how to train it on ”your” organism • Genemark-ES does it automatically Cegma can help De novo gene prediction • Pros: Can give you both exons and CDSs, fast • Cons: Very dependent on good training, as less ”real” data is used, accuracy can suffer Transcripts assembled from mapped RNA-seq reads • Bowtie/Tophat/Cufflinks/, PASA • Reads are mapped to the genome, and transcripts are assembled based on the distribution of the reads Cufflinks Cufflinks Transcripts assembled from mapped RNA-seq reads • Pros: Based on what is expressed, retains isoforms, not biased • Cons: Sensitive to ”dirt” in data, can create too many isoforms , does not include CDS annotations Assemble transcripts de novo followed by mapping Assemble transcripts de novo followed by mapping • Overlapping reads are assembled into transcripts, often without reference • Trinity is commonly used, but Velvet with Oases can also be used, or Newbler if you have 454 rna-seq • If the data is clean and the coverage is good enough, full length transcripts will be assembled. These can be mapped to the genome using BLAT etc. Mapped Trinity-assembled transcripts Assemble transcripts de novo followed by mapping • Pros: Isoforms are retained, UTRs are included, only based on evidence from the studied organism • Cons: Can be ”noisy” with some intronic or intergenic sequences retained, CDSs not annotated Lifting over annotations from a closely related genome • Kraken • Align the two genomes and then transfer annotations between aligned regions • Alignment is best done using Satsuma Lift-overs Lift-overs Lifting over annotations from a closely related genome • Pros: Works great for closely related organisms, or re-assemblies, uses synteny which helps in orthology determination and functional annotation • Cons: Not really an annotation, not based on the target organism, dependent on good alignment Protein alignments • Not really an annotation, but very useful in combination with other data • Exonerate often used, but Scipio is also good • Conserved, little noise Protein aligments using Scipio Or combine everything • Combine protein-alignments, aligned transcripts, ESTs, and de novo gene predictions into one single annotation • Maker is the best ”light-weight” choice, but there is also EVM. Ensembl pipeline needs considerable expertise and resources. Evidencemodeler Maker Transcript annotation • Here the transcript is already defined. The challenge is to find where the coding regions starts and stops • Transdecoder Transdecoder Transdecoder Transcript annotation • Here the transcript is already defined. The challenge is to find where the coding regions starts and stops • Transdecoder • Trinotate Trinotate 2) Find out what the regions do, what do they code for? • Blastbased • Functional annotation based on functional domains • Transfering names based on overlapping coordinates • Blast2GO rocks! Blast2GO KEGG-mapping Some take home messages • Annotations are never final and are not the absolute truth • Annotations depend a lot on the method used • If you intend to use RNA-seq for annotation, stranded RNA-seq is great Some take home messages • Annotations are never final and are not the absolute truth • Annotations depend a lot on the method used • Maker is currently the best light-weight tool to combine different types of data • If you intend to use RNA-seq for annotation, stranded RNA-seq is great • Get help! BILS annotation resource • • • • • Starting up this autumn Uses the Ensembl pipeline and/or Maker Delivers high quality annotations Also available for consultation Biosupport.se Biosupport.se









