RNA-seq
advertisement
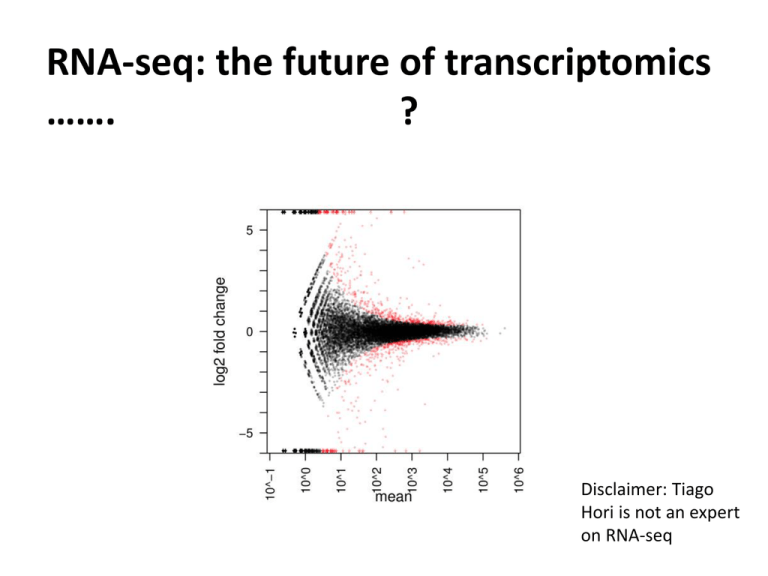
RNA-seq: the future of transcriptomics ……. ? Disclaimer: Tiago Hori is not an expert on RNA-seq Wang et al., 2009 • RNA-seq or RNA-sequencing is not a complete novel idea. • SAGE, long-SAGE, MPSS • The recent developments in next-generation sequencing (NGS) have made whole transcriptomic analyses more accessible. • Does it work? • Comparison with microarrays • Advantages and disadvantages • How does it work? • Challenges • Are microarrays going to go extinct? Weapons of choice: Marioni et al., 2008 There is a good correlation between microarray intensity and count data. There is also good correlation between Affymetrix foldchanges and Illumina-based RNA-seq fold-changes The Pros and Cons of RNA-seq – do the benefits definitely outweigh the problems? Advantages: • Allows for not only the identification of differentially expressed genes, but also identification of differential allelic expression, SNPs, splice variants, new genes or isoforms. • It is not limited to a set number of probes. • It is NOT impacted by background signal or saturation that causes problems in studying highand low-expression transcripts. Wang et al, 2009 The Pros and Cons of RNA-seq – do the benefits definitely outweigh the problems? Disadvantages: • Cost • Dependent on a reference genome or transcriptome. * see Trapnell et al., 2010 – Nature Biotechnology (used 430 million paired-end reads to assemble a transcriptome de-novo • Large amounts of data requiring large storage space and computational power • Statistical methods are still in their infancy How does it work? A) Agilent polyA selection B) NibleGen selection array C) Generation of target cDNA (sequence specific, e.g. for allele discrimination) D) Helicos sequencing Ozsolak and Milos, 2011 How does it work? Oshlack et al., 2010 Mapping Challenges: •Computational power required •Exon junctions •Alleles and SNPs Two main methods: Based on hash tables (local alignment similar to BLAST) Based on prefix/suffix trie BFAST Homer et al., 2009 BWA-SW Li and Durbin et al., 2010 One of the biggest challenge with mapping is to reduce the “RAM footprint” of the reference genome. This is accomplished by different ways of indexing the reference. The other challenge is to map accurately while allowing for variable reads (e.g. SNPs or error) to be mapped. Data summarization: There are 3 main ways of summarizing your data: 1. Counts per exon 2. Counts per transcript 3. Counts per gene (Oshlack et al., 2010) Normalization: Is RNA-seq data absolute mRNA count? • Within libraries: • Length bias • Sequencing efficiency • Between libraries: • Sequencing depth • Over-representation of highly-expressed transcripts Differential Expression detection: Challenges: • Requires biological replication but perhaps not technical replication. • Count data is discrete rather than continuous. • There is evidence the count data follow a negative binomial distribution similar to the Poisson distribution. • Accounting for type I error (False-Discovery) Bioconductor packages: edgeR: Developed for SAGE uses a modified Fisher exact test for dispersed data (means and variance estimated using maximum likelihood) DESeq: Similar to edgeR but uses a different model to estimate means and variance (empirical estimation of mean-variance relationship) BaySeq: Empirical Bayes inference to test of differential expression Systems Biology: DAVID and other microarray techniques used for GO enrichment KEGG pathways What do you do with data and what does it all mean? Resources: