Aligning reads with Galaxy
advertisement

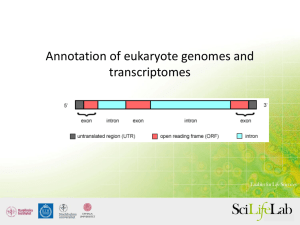
Finding genes de novo with RNA-seq Graham Etherington Graham.Etherington@tsl.ac.uk Today's topics • The basics – What is RNA-seq, alternative splicing. • Assembly techniques – Reference-based alignment – De-novo assembly • Expression analysis Today's topics • Tutorials in Galaxy – Finding genes through transcript assembly • TopHat – Cufflinks – Expression analysis • Cuffcompare – Cuffdiff RNA-seq – the basics • Genome of interest. – How many genes are there? – Are some novel? – Alternative spliced isoforms? – Are some transcripts more abundant than others? – Which genes are expressed under different environmental or biological conditions (e.g. lack of a nutrient, pathogen infection, etc)? What is RNA-seq? Genome Genes Extract mRNA (transcribed genes) Sequence RNA-seq basics - Alternative splicing Reference-based Alignment • Use when a closely-related reference is available. • 3 steps ① Use a splice-aware aligner (e.g. BLAT, TopHat) to align reads to a reference genome. ② Cluster reads from each locus to build isoform De Bruijn graphs. ③ Traverse graph to resolve isoforms. Each different path through graph represents a potentially different isoform. Alignment Seed and extend alignment (e.g. BLAST) Query ATCGCGTTACGATCCGTAA Find all occurrences of ‘ATCGCG’ ATCGCGGTCGTTAATCGCGCGTTCGATCGCGTTACGATCCGTAACGCACCATCGCGTTGC Seeds Target Alignment Seed and extend alignment (e.g. BLAST) Query ATCGCGTTACGATCCGTAA Extend alignments Genome ATCGCGTTAGTTAATCGCGTTACCGATCGCGTTACGATCCGTAACGCACCATCGCGTTAA Alignment • Burrow-Wheeler Transform (BWT) – used by BWA, SOAP, Bowtie (and TopHat) aligners • Creates a compressed index of the genome. • Index is a sorted range of substrings from genome that can be quickly searched. • Stretches of sequence can be looked-up – Like the index of a book. Words (sequences) can be looked up in index which then points you to the pages (genomic locations) were that word (sequence) is found. • Narrows-down the search space (searches index instead of genome) • Speeds up alignment and requires less memory when compared to older alignment algorithms. Creating and Traversing Graphs Aligned reads Create graph that represents alternative splicing Traverse graph to find all possible paths All possible splicevariants from graph Reference-based Alignment • Preferable where a high-quality reference exists. • Can assemble full-length transcripts at depth of 10x. • Advantages: – – – – Contamination not a great problem – won’t align. Less memory use than de novo assembly Detection of low-abundance transcripts Identify transcripts undiscovered in annotated reference Reference-based Alignment • Disadvantages: – Relies on the accuracy of the reference sequence • May contain errors, deletions, missassemblies. • Can miss divergent transcripts – Reads often align to multiple regions • Excluding multi-mapped reads – leaves gaps • Randomly assign multi-mapped reads – false transcripts – Can’t easily assemble trans-spliced genes (2 premRNAs spliced together to form 1 mature mRNA) De-novo assembly • Doesn’t use a reference sequence. • Constructs De Bruijn graph by breaking reads into k-mers and connecting overlapping nodes. • Graph is traversed to identify paths through it. • Each path represents a unique sequence. De Bruijn graphs • All substrings of length k (k-mers) are generated from each read. • 5-mers in this example De Bruijn graphs • • • • Overlapping k-mers used to create nodes in graph. Chains of adjacent nodes in graph are collapsed into a single node Alternative paths through graph are identified. Isoforms identified De-novo Assembly • Advantages – Doesn’t need a reference sequence. – Sometimes better than reference-based assembly when: • reference is of low quality (e.g. missing bits). • Unknown exogenous transcripts want to be detected. • Where long introns are expected. – Doesn’t depend on the correct alignment of reads to splice sites. De-novo Assembly • Disadvantages: – Lots of data requires lots of RAM – Requires greater sequencing depth than reference-based assembly (30x cf 10x). – Highly similar transcripts are likely to be assembled into single transcripts. – Sensitive to read-errors. Hard to tell errors from low-abundance transcripts. Expression analysis The more abundant an RNA, the more times it will be randomly selected for sequencing. The Cufflinks tool suite assembles transcripts and calculates their abundance. Sample 1 Gene A (control) Sample 2 Gene A (infected) expressed mRNA sequencing reads Expression analysis • Use number of mapped reads as an indicator of expression. Map reads back to genome Sample 1 Gene A (control) Differential expression Sample2 Gene A (infected) Normalisation • 2 sequence libraries can produce different volumes of data – transcript A present in same abundance in library X and library Y – library X produces 3 times more reads than library Y – transcript A in library X will appear 3 times more abundant. • Need some way to normalise the expression data. • Fragments Per Kilobase of exon, per Million fragments mapped (FPKM). – accounts for the number of reads in experiment, length of transcript and the number of reads aligning to it. – allows a comparisons between two datasets when there is considerably more data in one dataset than the other. Tutorials • Go through the tutorial sheet. • The task: – Reference-based RNA-seq assembly using TopHat and Cufflinks in Galaxy. – RNA-seq expression analysis using Cuffcompare and Cuffdiff in Galaxy. http://galaxy.tsl.ac.uk