Transcriptome analysis
advertisement
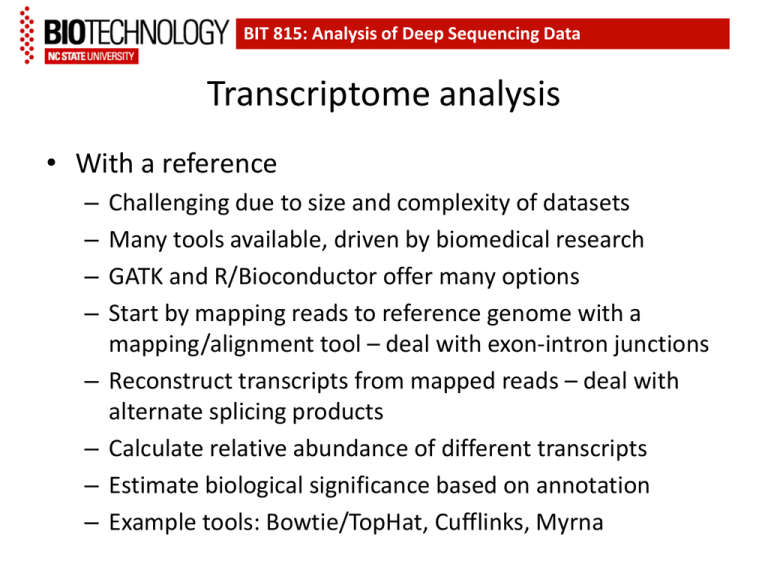
BIT 815: Analysis of Deep Sequencing Data Transcriptome analysis • With a reference – – – – – – – – Challenging due to size and complexity of datasets Many tools available, driven by biomedical research GATK and R/Bioconductor offer many options Start by mapping reads to reference genome with a mapping/alignment tool – deal with exon-intron junctions Reconstruct transcripts from mapped reads – deal with alternate splicing products Calculate relative abundance of different transcripts Estimate biological significance based on annotation Example tools: Bowtie/TopHat, Cufflinks, Myrna Workflow summary from a review “From RNA-seq reads to differential expression results”, by Oshlack et al, Genome Biol 11:220, 2010. Note emphasis on statistical analysis methods; an equal emphasis should be placed on experimental design. BIT 815: Analysis of Deep Sequencing Data The ‘Tuxedo’ suite of programs: Bowtie, TopHat, Cufflinks and CummeRbund See Trapnell et al, Nature Protocols 7:562 – 578, 2012 for details •TopHat maps reads •Cufflinks assembles transcripts •Cuffmerge merges transcript data detected in different treatments •Cuffdiff evaluates differential expression •CummeRbund provides visualization tools BIT 815: Analysis of Deep Sequencing Data Why merge data across treatments? BIT 815: Analysis of Deep Sequencing Data Differential transcript abundance mechanisms BIT 815: Analysis of Deep Sequencing Data Transcriptome analysis • Without a reference – First step is assembly – Transcriptome assembly pipelines • • • • Velvet/Oases – Oases is a post-assembly processor for Velvet Trans-ABySS (BCGSC) – based on ABySS parallel assembler Rnnotator – based on Velvet Trinity (Broad Institute) – a set of three programs – Common strategy: Assembly at multiple k-values, then merging of resulting contigs, followed by refinement – Once an assembly is available, continue with analysis as before BIT 815: Analysis of Deep Sequencing Data After Transcriptome Assembly… • Some amount of analysis of differential splicing versus differential promoter activity is possible, but conclusions may be less robust in the absence of a reference • The fraction of the total number of genes that can be discovered by RNA-seq depends on the diversity of tissue types and developmental stages analyzed, as well as the depth of sequencing 330 million SOLiD reads from a human cell line detect only about 67% of all annotated transcripts in the human genome. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Labaj et al, Bioinformatics 27:i383-91, 2011 BIT 815: Deep Sequencing Transcriptome analysis with RSEM RNA-Seq with Expectation Maximization Li & Dewey, BMC Bioinformatics 12:323, 2011 (a). Allows estimation of transcript abundance without a reference genome, based on alignments to assembled transcripts, although the transcripts can be taken from a reference genome sequence if it is available (b). Uses the Bowtie aligner by default, but considers reads that map to multiple locations in the reference transcript collection (c). For each sample, files of estimated transcript and isoform abundance are produced, along with SAM files of alignments. (d). The files of transcript and isoform abundance can be used to evaluate differential expression using tools from R and Bioconductor