gene (Pun1? - Plant Sciences
advertisement

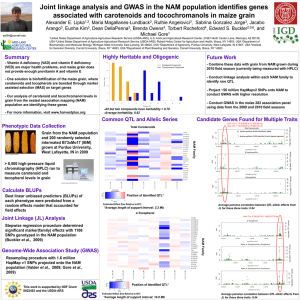
The genetics of heat –or pungencyin Capsicum Marco Hernandez-Bello PLS221 Introduction • Pungency or “heat” is due to accumulation of alkaloid capsaicin and its analogs in the placental tissue • Capsaicin biosynthesis is restricted to Capsicum – Driven domestication of several species • Has also an ecological role • Pepper species and cultivars differ with respect to their level of pungency (quantitative & qualitative) • Capsaicin has wide applications • Little is known about its biosynthesis (molecular, genetics, localization, accumulation) • Absence of pungency controlled by single recessive gene, pun1 C. annum C. chinense ‘Habanero’ C. frutescens ‘Tabasco’ www.thechileman.org/guide_species.php www.wikipedia.org The Pun1 gene for pungency in pepper encodes a putative acyltransferase Stewart, C. et al. 2005. Plant J. 42:675-688 Background • Little is known about the pungency accumulation • Absence of pungency controlled by pun1 – Single genetic source for non-pungency – single recessive gene – epistatic to all other pungency-related genes – Qualitative effect on presence/absence of capsaicinoids – Is it a master regulator of the pathway? – Mapped to chromosome 2 – cDNA from SSH library co-segregated with pungency in C.chinense – CAP marker has been used in breeding programs • Objective: Cloning and characterization of gene (Pun1?) responsible for pungency – Candidate gene approach Results Identification of SB2-66 as candidate gene for Pun1 - C. frutescens BG2816 (Pun1/Pun1) X C. annuum cv. Maor (pun1/pun1) Bell pepper - F2 mapping pop. (n=256) - cDNA SB-66 mapped to same region as Pun1 Results Identification of SB2-66 as candidate gene for Pun1 - Pungent (C. chinense, C. frutescens & C. annuum) and non-pungent (C. annuum) genotypes were surveyed to detect polymorphisms - Gene family? - Presence/absence band identified SB2-66 as candidate for Pun1 Results Characterization of cDNA and genomic sequence of Pun1 - Full cDNA and gDNA sequence from C. chinense ‘Habanero’ - C. annum ‘Thai Hot’ gene has 98% nt identity, and same structure - Conservation of deletion is widespread Results Sequence analysis - SMART predicted that AT3 has a acyltransferase domain (>40% similarity from other genes in plants), belongs to BAHD superfamily - AT1 & AT2 from Habanero fruit also showed high similarity with acyltransferases, but not mapped to Pun1 Results Regulation of AT3 expression - Habanero Pun1 and Bell pun1 peppers Results Regulation of AT3 expression - Expression during fruit development in C. annuum Pun1 and pun1 genotypes - Thai Hot is amenable to VIGS - Correspondence between transcript and protein accumulation - AT3 no detectable in Bell pepper - AT3 is tissue specific (only in placenta but pericarp & seeds) Results Function of AT3 in vivo by VIGS - Construct consisted of 400bp spanning active site - Agrobacterium-mediated transformation with tobacco rattle virus - Environmental stress may result with an increase in pungency? - Detection limits issues in inmmunoblot and HPLC - All the above provide evidence that Pun1 encodes an acyltransferase involved in capsaicinoid biosynthesis (is it the capsaicin synthase?) ? ? Results Phylogenetic analysis - 35 AT3 plant homologs are functionally characterized, and members of BAHD - AT3 falls in O-acetyltransferases (ester-forming enzymes) - AT2 likely fruit ripening/woundinginduced, closely related to N-acyltransferases Conclusions • Pun1 is an acyltransferase (AT3) involved in capsaicinoid biosynthesis – Capsaicin synthase? – CS is detected in non-pungent peppers – May be coA-dependent acyltransferase • pun1 originated trough a deletion and has been used for more than 300 years – Arose early in domestication? – Its use has narrowed genetic diversity? • AT3 activity remains to be elucidated – Mutants may identify accumulation of intermediates – Biochem assays remain challenging Characterization of capsaicin synthase and identification of its gene (csy1) for pungency factor capsaicin in pepper (Capsicum sp.) Prassad, B.C.N., et al. 2006. PNAS 103:1335-20 Background • Capsaicin is biosynthesized by CS – Condensation of vanillylamine and fatty acids moieties in placenta • Role of intermediates (8-methyl nonenoic acid) in capsaicin biosynthesis • Biotransformation of phenyl propanoid intermediates to capsaicin has been demonstrated • No reports of purification and cloning of CS gene • Objective: Identify the gene responsible for capsaicin biosynthesis – Enzyme-to-gene approach Results Purification of CS - Correlation between CS activity and pungency - CS from high pungency genotype was purified and characterized Results Purification of CS - Crude placental protein was extracted - 110 fractions obtained, CS assayed and bulked - Purification was enhanced by Sepharose column with bound vanillylamine Results Expression of CS during fruit development - Polyclonal antibodies were highly specific to CS - CS and capsaicin levels are correlated - CS is localized in peripherial cells of placental tissues High pungent C. frutescens 7d 15d 14 22 21 28 35 30 42 L/M C. annuum 50 28 45 28 Results Identification of CS gene - N-terminal amino acid sequence was determined - Primers were design to amplify a N-terminal motif –rev primer from SB2-66 - Gene is 981 bp, has no introns, 308 aa and predicted 38 kDa molecular mass - NO significant homology with any reported amino acid sequences (including acyltransferases) cDNA gDNA Results Expression of csy1 - csy1 expressed only in placenta - Transcript level correlated with pungency genotypes - Sequences from C. frutescens & C. annuum were similar Results Heterologous expression of csy1 - csy1 was expressed in E. coli DH5a using pRESTA vector - CS showed higher specific activity than native CS - CS highly specific to substrates of CS - csy1 function is specific to capsaicin biosynthesis 35 kDa Conclusions • High pungency level correlated with high levels of capsaicin and CS activity • CS confined to peripherial cells of placental tissue • Levels of capsaicin and CS activity depends on genotype • csy1 is unique to Capsicum? QTL analysis for capsaicinoid content in Capsicum Ben-Chaim, A., et al. 2006. TAG 113:1481-90 Background • Presence/absence of capsaicinoid due to pun1 • Amount of capsaicinoid is a quantitative trait – Varieties with different levels of pungency – Pungency level is also affected by environment • Limited information on quantitative variation – cap, major QTL on chromosome 7 (C. frutescens x C. annuum) F2 – No co-localization between predicted structural genes and variation in capsaicinoid content – cap is a regulator of the pathway or unknown structural gene? • Objective: Identify genomic regions that may independently control presence of single capsaincinoid analogs – Using F2 & F3 pop’s C. frutescens BG2814-6 x C. annuum RNaky Results Linkage map construction - 728 markers (SSR, AFLP, specific PCR-markers, RFLP), and candidate genes involved in capsaicinoid biosynthesis (pAMT, COMT, Bcat) - 12 major and 4 small linkage groups, total length 1358.7 cM Results Phenotypic variation and correlation among traits - Capsaicinoids in BG2814-6 (small fruit) was 10-30X-fold than RNaky (large fruit) - Content in F1 was higher than BG2814-6 parent (overdominance/heterosis) - F3 families showed transgressive segregation (except fruit weight) - Capsaicin was the most abundant (38-64%), nordihydro- the least - Capsaicin highly correlated w/ dihydro-, and moderately w/ nordihydro- - Capsaicin affected by environment, nordihydro- not Results QTL ID: capsaicin content - Alleles from pungent parent contributed to increased capsaicin content - cap7.2 has large QTL effect (>20%) - Both additive (cap3.1, cap4.1) and dominant (cap7.1) gene action - Phenotypic variation by all QTL was 24, 19, and 37% in 2001, 02 & 03 - Digenic interaction detected between cap7.1 and marker in chr 2 (NP0326) Results QTL ID: dihydrocapsaicin content - Four of the 5 QTL for capsaicin, were also identified - Alleles from pungent parent contributed to increased dihydrocapsaicin content - Same digenic interaction as befor Results QTL ID: digenic interaction - Presence of BG2814-6 alleles at both positions was correlated with the largest increase of capsaicin (37-42%) and dihydrocapsaicin contents (24-28%) Results QTL ID: nordihydrocapsaicin/total content, fruitweight - Ndhc7a.1 didn’t co-localize with QTL controlling other capsaicinoids and was recessive - 5 QTL for total capsaicinoid content, total7.2 has the largest effect - 2 QTL for fruit weight, gene action dominant (fw2.1) and additive (fw3.1) Results Co-segregation of candidate genes w/QTL - Genes 3A2 and BCAT (valine catabolism) co-localize with QTL - 3A2 has motifs of hydroxyisobutyrate dehidrogenase - BCAT involved in catabolism of branched-chain amino acids Conclusions • Identified QTL may represent elements in pathway • 2 independent QTL found – cap3.1 influenced capsaicin and total capsaicinoid content – ndhc7a.1 affected only nordihydrocapsaicin • Overlapping QTL suggests common genetic mechanisms • Fruit weight QTL didn’t co-localize w/ capsaicinoid QTL • cap7.2 likely orthologous to major QTL previously identified • Digenic interaction may facilitate further genetic analysis