RIBOZYMES
advertisement

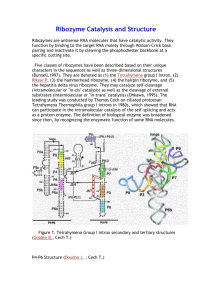
ASHA S V BCH-10-05-05 Ribozyme, or RNA enzyme, is a RNA molecule that act as enzymes, often found to catalyze cleavage of either its own or other RNAs. Due to their complex secondary structures and hairpin/hammer head active centres, RNAs could act as a catalyst and this idea was proposed by Carl Woese et al, Thomas R Cech and Sidney Altman were the first to discover ribozyme. It has also been found to catalyze the aminotransferase activity of the ribosome. The RNA catalysts called ribozymes are found in the nucleus, mitochondria, and chloroplasts of eukaryotic organisms. Some viruses, including several bacterial viruses, also have ribozymes. Almost all ribozymes are involved in processing RNA. They act either as molecular scissors to cleave precursor RNA chains (the chains that form the basis of a new RNA chain) or as "molecular staplers" that ligate two RNA molecules together. Although most ribozyme targets are RNA, there is now very strong evidence that the linkage of amino acids into proteins, which occurs at the ribosome during translation, is also catalyzed by RNA. Thus, the ribosomal RNA is itself also a ribozyme. Types of ribozymes Ribozymes may be classified into natural ribozymes and artificial ribozymes Natural ribozymes include, Peptidyl transferase 23S rRNA,RNase P, Group I and Group II introns, G1R1 branching ribozyme,Leadzyme, Hairpin ribozyme, Hammerhead ribozyme, HDV ribozyme, Mammalian CPEB3 ribozyme, VS ribozyme, glmS ribozyme, CoTC ribozyme Artificial ribozymes are synthesised in the laboratory based on the dual nature of RNAs as a catalyst and an informational polymer . Synthesis of artificial ribozymes involves the mutation of natural ribozymes. This process is initiated by reverse transcription using reverse transcriptase. this results in the generation of various cDNA. Relatively short RNA molecules that can duplicates others have been artificially produced in the lab. This includes a 165-base long RNA and a 189-base long RNA. these ribozymes could polymerize RNA primers. A short RNA molecule which mimics one of the two rRNAs of ribozymes has been synthesised invitro. this short strand of rRNA catalyzes the synthesis of peptide bond. Hammerhead ribozyme Hammerhead RNAs are RNAs that self-cleave via a small conserved secondary structural motif termed a hammerhead because of its shape. Most hammerhead RNAs are subsets of two classes of plant pathogenic RNAs: the satellite RNAs of RNA viruses and the viroids. The minimal hammerhead ribozymes The minimal hammerhead sequence that is required for the self-cleavage reaction includes approximately 13 conserved or invariant "core" nucleotides, most of which are not involved in forming canonical Watson-Crick basepairs. The core region is flanked by Stems I, II and III, which are in general made of canonical Watson-Crick base-pairs but are otherwise not constrained with respect to sequence. The catalytic turnover rate of minimal hammerhead ribozymes is ~ 1/min . Much of the experimental work carried out on hammerhead ribozymes has used a minimal construct. The crystal structure of a minimal hammerhead ribozyme Structurally the hammerhead ribozyme is composed of three base paired helices, separated by short linkers of conserved sequences. These helices are called I, II and III. Hammerhead ribozymes can be classified into three types based on which helix the 5' and 3' ends are found in. If the 5' and 3' ends of the sequence contribute to stem I then it is a type I hammerhead ribozyme, and if the and 3' ends of the sequence contribute to stem III then it is a type III hammerhead ribozyme. Of the three possible topological types both type I and type III are common. It is not known if examples of the type II topology are found in nature. Hammerhead ribozyme (type I) Hammerhead ribozyme (type III) The full-length hammerhead ribozyme The full-length hammerhead ribozyme consists of additional sequence elements in stems I and II that permit additional tertiary contacts to form. The tertiary interactions stabilize the active conformation of the ribozyme, resulting in cleavage rates up to 1000-fold greater than those for corresponding minimal hammerhead sequences. Three-dimensional structure of the full-length hammerhead ribozyme Hairpin ribozyme The hairpin ribozyme is a small section of RNA that can act as an enzyme known as a ribozyme. Like the hammerhead ribozyme it is found in RNA satellites of plant viruses. It was first identified in the minus strand of the tobacco ring spot virus (TRSV) satellite RNA where it catalyzes self-cleavage and joining (ligation) reactions to process the products of rolling circle virus replication into linear and circular satellite RNA molecules. The hairpin ribozyme is similar to the hammerhead ribozyme in that it does not require a metal ion for the reaction Leadzyme Leadzyme is a small ribozyme that was artificially made using in vitro selection techniques. Leadzyme is able to cleave RNA in the presence of lead The structure of leadzyme has been determined by Xray crystallography It has been proposed that a naturally occurring leadzyme occurs in the 5S rRNA and further that this may be an important mechanism in lead toxicity. Hepatitis delta virus ribozyme HDV ribozyme is composed of five helical segments connected by a double pseudoknot. Pseudoknot is a kind of teritiary interaction in RNA which can join distant parts of the same RNA strand. It is involved in a self cleavage process needed for processing the RNA transcripts and it is needeD for viral replication. HDV ribozyme is the fastest known natural self cleaving RNA. The Varkud satellite (VS) ribozyme The Varkud satellite (VS) ribozyme is an RNA enzyme that carries out the cleavage of a phosphodiester bond. The VS ribozyme is composed of 5 helices that form an H shape (helices II to VI). The first helix (I) contains the substrate cleavage site in the stem-loop Reactions catalyzed by ribozymes Ribozymes can catalyze a whole range of reactions This includes hydrolysis of their own RNA component by cleaving the phosphodiester bond[these ribozymes are called RNA polymerizing ribozymes] as well as hydrolysis of other RNA strands and aminotransferase activity of the ribozyme. A list of rections catalysed by ribozymes are; Hydrolysis of other RNA strands Catalysis of self synthesis by templated polymerization Self splicing DNA and RNA phosphorylation by synthetic ribozyme Catalysis of peptidyl transferase reaction during translation Aminotransferase reaction of the ribosome Catalysis of tRNA maturation Catalysis of self ligation and self cleavage Chaperon reactions Ribozymes as a cofactors RNA can act as a cofactor for amino acid residues. Amino acid complexes with RNA molecules during which the later functions as a cofactor, enhancing or diversifying the enzymatic capabilities of proteins. mRNAs have evolved from RNA molecules which catalyzes amino acid transfer to them Chaperon like ribozymes Like chaperon, RNA catalyzes protein folding. Such RNAs are called chaperon like ribozymes Applications A type of synthetic ribozyme directed against HIV RNA called gene shears has been developed and has entered clinical testing for HIV infection reference Enzymology ;T Devasena Functional metabolism regulation and adaptation;Kenneth B Storey Text book of biochemistry ;Thomas M Devlin