Ribozyme Catalysis & Structure: Types, Function, & Mechanisms
advertisement

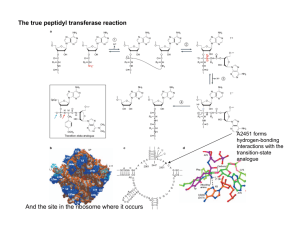
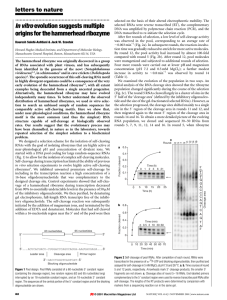
Ribozyme Catalysis and Structure Ribozymes are antisense RNA molecules that have catalytic activity. They function by binding to the target RNA moiety through Watson-Crick base pairing and inactivate it by cleaving the phosphodiester backbone at a specific cutting site. Five classes of ribozymes have been described based on their unique characters in the sequences as well as three-dimensional structures (Bunnell,1997). They are denoted as (1) the Tetrahymena group I intron, (2) RNase P, (3) the hammerhead ribozyme, (4) the hairpin ribozyme, and (5) the hepatitis delta virus ribozyme. They may catalyze self-cleavage (intramolecular or "in-cis" catalysis) as well as the cleavage of external substrates (intermolecular or "in-trans" catalysis) (Ohkawa, 1995). The leading study was conducted by Thomas Cech on ciliated protozoan Tetrahymena Thermophila group I introns in 1980s, which showed that RNA can participate in the intramolecular catalysis of the self-splicing and acts as a protein enzyme. The definition of biological enzyme was broadened since then, by recognizing the enzymatic function of some RNA molecules. Figure 1. Tetrahymena Group I intron secondary and tertiary structures (Golden B., Cech T.) P4-P6 Structure (Doudna J. , Cech T.) Figure 2. A hammerhead ribozyme structure with magnesium cation binding sites (Murray,1996) The name of hammerhead ribozyme is given by the similarity between its secondary structure and the shape of a hammerhead. They are the best understood subcategory of all ribozymes. As well as other ribozymes, the hammerhead ribozyme is an antisense RNA. Some of the ribonucleotides within the sequence selectively form Watson-Crick base pairs with others to form a stem, while the rest stay in single stranded state called loop. These loops and stems can be predicted at the secondary structure level using conformational energy analysis, such as RNAdraw and mfold; and three dimensional structures were obtained mainly by X-ray crystallography. Figure 3. Secondary structures of in cis and in trans hammerhead ribozymes On the left of Figure 3, the structure of the wild-type, cis-acting hammerhead ribozyme is shown. The three helices and the cleavage site are indicated. The self-splicing happens after the sequence G-U-C between helix I and III, and the G and U are conserved and crucial for the catalysis. Except for itself, a hammerhead ribozyme is not catalytic in cells since its nature of intramolecular reaction (Symons, 1989). However, most ribozymes can be chemically engineered to cleave RNA in trans by separating the catalytic domain from ribozyme and attaching recognition (i.e., antisense) arms to the catalytic center, in order to target to a substrate. These trans-acting ribozymes can be very useful in the study of molecular biology and pharmaceutics. In comparison to the conventional antisense RNAs, ribozymes provide the potential of turnover, with a single molecule being able to inactivate multiple target RNAs. Another important property of ribozyme is the specificity, Figure 4. Turnover cycle of RNA cleavage by a hammerhead ribozyme. Binding of the enzyme and substrate results in the catalytically active structure. After cleavage, the two product strands dissociate and the ribozyme strand can go into the next cycle. which means the fidelity to cleave a unique target. Both of turnover and specificity are affected by binding arm length (helix I and III). If the length of binding arms is very short, the rate of dissociation of the target from the ribozyme may exceed the rate of cleavage, resulting in poor efficiency (Rossi, 1997). However, stable hybrids exhibit low catalytic activity because of slow dissociation of the cleaved substrate (Bertrand, 1994). Thus, the ideal situation is to have arm lengths that aid cleavage, yet provide for quick dissociation of the cleaved products. This present web page is part of a term paper assignment for the Advanced Enzymology Class (Quan Du).