rna evolution by recombination and selection
advertisement

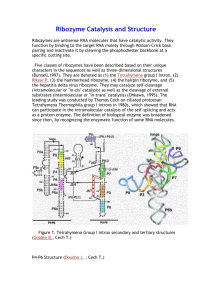
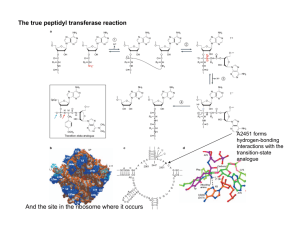
Ribozymes Evolution by Random Recombination and Selection Qing (Sunny) Wang and Peter J. Unrau Molecular Biology and Biochemistry Department, Simon Fraser University Burnaby, B.C. Canada V5A 1S6 HYPERLINK "mailto:qwangb@sfu.ca" qwangb@sfu.ca, HYPERLINK "mailto:punrau@sfu.ca" punrau@sfu.ca Ribozymes (catalytic RNAs), which combine both genotype and phenotype into a single molecule, are good models to study the role of recombination in the evolution of the ‘RNA World’. More and more natural and artificially selected ribozymes were found to be with modular structure, in which sequence that are essential for function are separated from flanking regions that are required to form a continuous strand of RNA. This modularity indicates the great potential for nonhomologous recombination to evolve new functional ribozymes efficiently. We have recently shown that nonhomologous recombination is a highly efficient way to isolate a ribozyme’s core motif by removing nonessential sequence that is present in the original ribozyme sequence. Approximately 108 scrambled RNA variants were constructed by randomly recombining fragments of a 271 nucleotide long pyrimidine synthase ribozyme. In vitro selection quickly resulted in the isolation of a diverse size range of ribozyme variants that were as active as their much longer progenitor. The shortest ribozyme, only 81 nucleotide long, consisted of four regions of sequence that were conserved by all of the truncated active variants and therefore represents the core motif sequence. Using a similar methodology, we are now attempting to isolate RNA-ligase ribozymes from a population of randomly recombined RNA fragments derived from several nucleotide synthase ribozymes. If successful, this will be the first time demonstrating that new ribozyme can evolve by recombining fragments of functional unrelated ribozymes. Analyzing the origin of the fragments appearing in the new functional ribozymes may help us explore the following fundamental questions: What criteria are required for RNA molecules to build up new functions from old? Biophysics Bash 2005 Qing (Sunny) Wang Simon Fraser University, B.C., Canada