Finding TFBS
advertisement

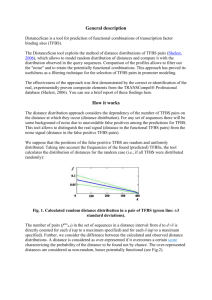
Finding Transcription Factor Binding Sites BNFO 602/691 Biological Sequence Analysis Mark Reimers, VIPBG Questions • If know motif (or sequence binding preferences) can you identify likely active TFBS? • If you have a TF, can you find its motif and binding sites? • Can you find motifs and binding sites for unknown TFs? Finding TFBS and Motifs in Animals • Sequence-based methods – If know sequence, scan known TFBS motif across genome • Data-based methods – Use ChIP to identify locations of binding • Needs good antibody; often picks up indirect binding – Compare promoters across genomes • Need depth; miss enhancers and species-related changes – Look for DNAse footprints – Use SELEX or DS-DNA microarray to profile TF’s DBD • Ideally combine both kinds of methods Outline • Bioinformatics approaches: PSWM • Experimental approaches to finding TFBS • Integrated approaches Position-Specific Weight Matrices Represent TFBS Better than Motifs • Represent log of probability of each base occurring at each position in TFBS • Often used to scan along genome calculating log-likelihood at each position A composite PWSM scan for SP1 (from PEAKS webpage) Standard Scoring Form of PSWM • Goal to compute probability of sequence relative distribution on sets of sequences bound by TF, compared to probability under random distribution • Assume independence of bases to simplify – Not bad for many; bad for some • Log likelihood of sequence would be sum of LL for base i in position j: log2(pij / bi) – pij is proportion of occurrences of base i – bi is baseline proportion of base i • If bis differ a lot from uniform then independence assumption often invalid – Many false positives from scan Experimental Approaches to Identifying TFBS and Motifs ChIP-Seq Can Identify Many TFBS • Chromatin Immuno-precipitation can identify where a TF binds to the genome • One can try to identify sequences that occur more often than chance by a variety of methods • Caveat: indirect binding may have wrong motif From Rozowsky et al, Nature Biotech 2009 Other Approaches to Finding TFBS • Systematic Evolution of Ligands by Exponential Enrichment (SELEX) From Jolma et al, Cell, 2013 Generate random DNA sequence library of moderate length. The sequences in the library are exposed to the target ligand, and those that do not bind the target are removed by affinity chromatography. The bound sequences are eluted, and then amplified by PCR, and the process is run again under more stringent elution conditions to purify the tightest-binding sequences. Finding TFBS by DNase Footprints From Neph et al, Nature, 2012 Identifying TFBS by Novel Recurrent Motifs under DNaseI Footprints From Neph et al, Nature, 2012 Integrated Approaches to Identifying Active TFBS in Tissues Integrated Approaches to Identifying TFBS • In this course we focus on binding sites for transcription factors with known motifs • Combining PWM Scores and other genomic data – PhastCons or PhyloP conservation – DNAse and histone marks – Integrating DGF • We will combine information using a Bayesian framework Bayesian Hierarchical Model for Integrating Information PSWM Score distributions Prior Probability of TFBS Posterior probabilities Conservation distribution DNase distribution Bayesian Hierarchical Models • Prior probability of binding site set very low or estimated from TF-specific ChIP data • In principle binding should be a continuous variable; we will treat as ‘yes-no’ • Need to estimate probability of various genomic features – conservation, DNAse – for TFBS and for background sequence What Information from Histone Marks? • By themselves histone marks, esp H3K4me3, H3K4me1, H3K27me3 can be very informative • After introducing DNAse data, these marks do not add much direct information • Could be used to adjust probabilities for DHS and conservation (not yet done) Bayes Model for Combining PWM Scores and Conservation • How to estimate P(conserved | TFBS)? • Depends on depth of time for which conservation is used – For mammals ~ 40%; primates ~ 80% – Varies between promoter and enhancer • Background state can be estimated from genome-wide conservation (typically 5 - 10%) • Then combine by Bayes Formula P(B | C, S) = P(C & S | B)P(B) P(C & S) P(C & S | B)P(B) = P(C & S | B)P(B) + P(C & S | ~ B)P(~ B) • C and S are conditionally independent given B, so P(C&S|B) = P(C|B)P(S|B) (likewise for ~B) Bayes Model for Combining Scores and DNase Sensitivity • How to estimate P(DHS | TFBS)? • Almost all (~98%) of known TFBS occur in DHS • Background state can be estimated from genomewide levels (typically 1 or 2%) • Then combine by Bayes Formula P(D & S | B)P(B) P(B | D, S) = P(D & S) P(D & S | B)P(B) = P(D & S | B)P(B) + P(D & S | ~ B)P(~ B) • D & S are conditionally independent given B, so P(D&S|B) = P(D|B)P(S|B) Chromia – A Method for Using Histone Marks and PSWM • Uses an HMM approach to integrate PSWM and histone marks (P300 marks enhancers) CENTIPEDE– A Method for Combining DNAse, Conservation and PSWM Scores • Combines several kinds of genomic information with PSWM to identify putative TFBS • Confirmation by ChIPSeq is quite good Pique-Regi R et al. Genome Res. 2011;21:447-455 CENTIPEDE– A Method for Combining DNAse, Conservation and PSWM Scores Model learned by the CENTIPEDE approach for the transcription factor NRSF. (A) Empirica density plots for key aspects of the data for sites inferred by CENTIPEDE to be bound (gree lines, CENTIPEDE posterior probabilities >0.95) and unbound (red lines, probabilities < 0.5 Pique-Regi R et al. Genome Res. 2011;21:447-455