Transcription Factor binding site analysis
advertisement

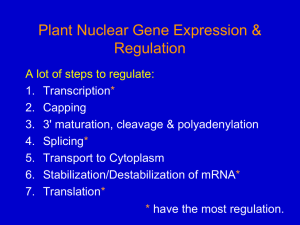
Promoter Analysis TFBS Detection Daniel Rico, PhD. drico@cnio.es 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR 2 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. TFBS prediction using PWMs 5. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR Transcription Factor Binding Sites 3 Enhancer Gene “Proximal” promoter (100bp-2Kb 5’ Upstream) TSS: Transcription Start Site 4 PROMOTERS Promoters are DNA segments upstream of transcripts that initiate transcription Promoter 5’ 3’ Promoter attracts RNA Polymerase to the transcription start site 5 GENES IN ENSEMBL 5’ Forward (+) strand 3’ Reverse (-) strand 6 Transcription Start Site Transcription Termination Site 7 PROMOTER STRUCTURE IN PROKARYOTES (E.COLI) Transcription starts at offset 0. • Pribnow Box (-10) • Gilbert Box (-30) • Ribosomal Binding Site (+10) 8 PROMOTER STRUCTURE IN EUKARYOTES 9 Experimental Transcription Start Sites (TSS) by CAGE CAGE (Cap Analysis of Gene Expression)) detects the transcriptional activity of each promoter transcript. 10 Representation of CAGE preparation protocol adapted to various platforms. Now Solexa and Illumina are preferred. 454 Life Sciences (FLX system) is not used any longer because concatenation requires additional PCR cycles and complicated manipulation. In the future, singlemolecule sequencing technology will be preferred because PCR may not be required. 11 http://www.osc.riken.jp/english/activity/cage/basic/ 12 http://fantom.gsc.riken.jp/4/edgeexpress/view/ 13 http://www.epd.isb-sib.ch/ 14 15 SEQUENCE ANALYSIS: SEARCHING TRANSCRIPTION FACTOR BINDING SITES (TFBS) 16 TFBS: DETECTION METHODS in vivo Functional analysis ChIP in vitro on cloned fragment Footprinting reactions Exonuclease digests Gel retardation (EMSA) UV Crosslinking in vitro on artificial DNA: SELEX: Systematic Evolution of Ligands by Exponential enrichment 17 TRANSCRIPTION FACTORS BIND TO TFBS IN DNA Affinity Specificity Nat Rev Genet. 2010 Nov;11(11):751-60. Epub 2010 Sep 28. Determining the specificity of protein-DNA interactions. 18 TF BINDING SITES Problems: often poorly defined consensus Sequences not conserved within species, and even worse between species Examples of enhancers functionally conserved but not sequence-conserved Most of the TFBS sequence data comes from just a few species Very often in vitro experiments 2 completely different binding sites could be merged in the same matrix/consensus 1 9 19 Transcription Factor Binding Sites 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR 20 Data collection Probabilities can be calculated and corrected for background Also called position-specific scoring matrices (PSSMs). In log scale. 21 FROM PFM TO PWM/PSSM Transcription Factor Binding Sites 22 SEQUENCE LOGOS: The information content of a matrix column ranges from 0 (no base preference) and 2 (only 1 base used). http://weblogo.berkeley.edu/ 23 http://www.lecb.ncifcrf.gov/~toms/sequencelogo.html Summary AAGTTC AAGCTC AGGCTC AAGGTC A C G T 430000 000204 014100 000140 Consensus: ARGBTC 24 Transcription Factor Binding Sites 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Obtain mouse and human fosB promoters and predict TFBS with Match and JASPAR 25 Transfac: not free, 848 matrices, loads of information and references, quality score based on methods used Jaspar: open sources, 123 matrices, minimal information, majority based on SELEX method (80%) 2 6 26 TRANSFAC® http://www.gene-regulation.com/pub/databases.html 27 http://jaspar.cgb.ki.se/ http://jaspar.genereg.net/ 28 JASPAR EXAMPLE: PAX6 29 2 9 Transcription Factor Binding Sites 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. Pattern Matching: TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR Transcription Factor Binding Sites 30 Click here to select all TFBS 31 Transcription Factor Binding Sites 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. Pattern Matching: TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR 32 PATTERN DISCOVERY Reference Genome Sequences of interest Seq. oligo expected frequency Seq. oligo observed frequency AAAAAA AAAAAC AAAAAG AAAAAT 0.00024 AAAACC … AAAAAA AAAAAC AAAAAG AAAAAT 0.00018 AAAACC … 0.00024 0.00030 0.00031 0.00028 0.00023 0.00031 0.00125 *** 0.00026 3 3 33 http://meme.sdsc.edu/meme/ 34 1. Promoters and gene regulation in Eukaryotes 2. Position Weight Matrices (PWM) 3. PWM Databases 4. Pattern Matching: TFBS prediction using PWMs 5. Pattern Discovery: Finding unknown motifs 6. Exercise: Use the human NOS2 sequence to predict TFBS with Match and JASPAR Transcription Factor Binding Sites 35 EXERCISE Step by step a. Download from UCSC or Ensembl the human NOS2 gene plus 5000 bases upstream. Select the “proximal promoter” first 1Kb: from -1000 to TSS (hint: there is no zero position!) b. Go to JASPAR and search for TFBS in promoter with the defaults. c. Do the same exercise with the mouse NOS2. d. Compare the results. 36 CHROMATIN ACCESSIBILITY 37 Access to experimental information http://www.nature.com/scitable/ EUCROMATINA Y HETEROCROMATINA Replicatión temprana (early) Replicatión tardía (late) Nat Rev Genet. 2011 Jul 12;12(8):554-64. doi: 10.1038/nrg3017. Determinants and dynamics of genome accessibility. Slides from http://www.openhelix.com/ENCODE ENCODE: WWW.GENOME.GOV/10005107 ENCyclopedia of DNA Elements, NHGRI Consortium of international researchers UCSC is the Data Coordination Center 47 ENCODE BACKGROUND Pilot phase, or phase I: www.genome.gov/26525202 Selected regions of the genome: 1%, 30 MB 48 ENCODE PILOT DATA AND BEYOND ENCODE portal: http://genome.ucsc.edu/ENCODE/ Pilot ENCODE browser: genome.ucsc.edu/ENCODE/pilot.html 49 ENCODE NEXT PHASE: PRODUCTION PHASE UCSC is the DCC for human and mouse data The portal is available: genome.ucsc.edu/ENCODE/ New aspects of the Production Phase projects 50 chromatin transcriptome/ genes Copyright OpenHelix. No use or reproduction without express written consent ENCODE PRODUCTION PHASE FOCUS promoters/ regulatory sites DNase sites ENCODE is now genome-wide Specific cell types and new technologies being applied Project focus topics selected, then supplemented 51 Data being submitted to UCSC DCC by data providers “Wranglers” ensure meta data is present Quality checks occur, data is released for use Copyright OpenHelix. No use or reproduction without express written consent ENCODE DATA IS FLOWING! 52 ENCODE DATA TYPES ENCODE Tracks identified with icon Mapping data Genes Expression Regulation Variation 53 REGULATION DATA Image from NIH Regulation data Structure: modifications, open vs. closed chromatin 54 REGULATION DATA II TATA bound to DNA Transcription factor binding sites, TFBS RNA binding proteins 55