M. tuberculosis - El Corte Inglés
advertisement

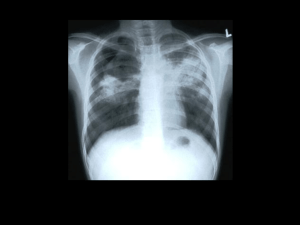
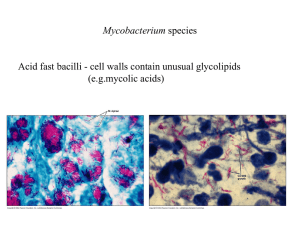
Mycobacterium tuberculosis Evolution of Functional Diversity Douglas Young A new horizon for preventive vaccines against tuberculosis Madrid 7th May 2014 Field trial of BCG in badgers Gloucestershire 2005-2009 844 badgers caught and sampled disease detection by serology 262 captured more than once were test negative on initial capture 22 incident cases 74% reduction in seropositive disease group no of badgers incident cases % of total cases CI control 82 14 17.1 (10.8-25.9) vaccinated 179 8 4.5 (2.4-8.2) F probability unvaccinated cubs from vaccinated setts had a reduced ESAT6/CFP10 IFNg response 0.001 79% reduction in IFNg conversion vaccination interrupts onward transmission Chambers et al. 2011. Proc Biol Sci B. 278:1913-20 Carter et al. 2012. PLoS One 7:e49833 Bovine TB in Ethiopia A. bovine TB in rural cattle 30000 carcasses screened in abattoirs 1500 lesioned animals, 170 ZN+ cultures low prevalence 0.5 – 5% 58 M. bovis isolates 8 M. tuberculosis isolates (12%) B. bovine TB in urban intensive farms high prevalence > 50% post-mortem: 67 cultures from 31 animals 67 M. bovis isolates 0 M. tuberculosis isolates M. tuberculosis can cause disease in individual animals, but it doesn’t establish an efficient transmission cycle Berg et al. 2009. PLoS One 4:e5068 Firdessa et al. 2012. PLoS One 7:e52851 THE CONCEPT I want to have a vaccine that interrupts transmission: can I target some layer of species-specific biology that is required for an effective transmission cycle? THE MODEL the ideal vaccine candidate biology involved in effective transmission biology involved in making a lesion THE STRATEGY I don’t have an experimental model for transmission, so I’m going to try and infer biology by looking at evolution of human isolates Global phylogeny of M. tuberculosis Lineage 7 Lineage 4 Lineage 1 Lineage 3 Lineage 5 Lineage 2 Lineage 6 Comas et al. 2013. Nat Genet 45:1176 animal strains Do toxin-antitoxin modules regulate “persistence”? transcription higher in Lineage 1 transcription higher in Lineage 2 Rose et al. 2013. Genome Biol Evol 5:1849-62 in vitro transcription profiling reveals strain variation in transcript abundance but there’s very little evidence of genomic diversity of TA modules Number of TA modules 0 10 20 30 40 50 60 70 80 M. tuberculosis M. canettii 60008 M. canettii 70010 Mycobacterium sp. JDM601 M. gastri M. kansasii M. xenopi M. yongonense M. paratuberculosis M. smegmatis mc2 155 M. avium M. marinum M. abscessus M. ulcerans M. phlei M. hassiacum Mycobacterium sp. MCS M. gilvum M. smegmatis JS623 M. chubuense blue: chromosome red: plasmid TAs and phylogeny high TA mycobacteria (>10 modules) in red 100 88 M. paratuberculosis deletion of lon protease M. yongonense 65 rpoC sequence, GTR+G+I, Maximum Likelihood phylogeny, 100 bootstrap M avium M. kansasii 76 100 M. gastri M. ulcerans 79 100 M. marinum ddn nitroreductase ddn nitroreductase lactate dehydrogenase lon protease lactate dehydrogenase M. canettii 70010 90 100 99 100 ddn nitroreductase M. tuberculosis M. canettii 60008 M. xenopi Mycobacterium sp. JDM601 62 M. phlei 96 M. hassiacum M. smegmatis JS623 57 M. chubuense 100 100 M. gilvum Mycobacterium sp. MCS 100 M. smegmatis MC2 155 M. abscessus 0.02 plasmids lactate dehydrogenase What else is carried on mycobacterial plasmids? toxin-antitoxin modules metal ion detox and efflux cytochrome P450s adenylate cyclases diguanylate cyclases Type VII secretion loci mce loci ... organism adenylate cyclase domains M. tuberculosis 16 M. marinum 31 M. ulcerans 15 M. smegmatis mc2 155 7 M. smegmatis JS623 48 ESX locus on pMK12478 MKAN_ chromosome 00155 56% MKAN_ plasmid 29475 57% 00160 00195 53% 91% 29470 29465 29460 PE PPE 52% Mtb Rv1783 Rv1784 eccB5 eccC5 00200 00205 00210 00215 00220 00225 95% 45% 50% 55% 34% 72% 29455 29450 29445 29435 29430 29425 29420 pseudo 94% 45% 48% 57% 31% 72% Rv1792 Rv1793 Rv1794 Rv1795 Rv1796 Rv1797 Rv1798 esxM eccD5 esxN Rv1785 Rv1786 Rv1787 Rv1788 Rv1789 Rv1790 Rv1791 cyp143 PPE25 PE18 29440 PPE26 PPE27 PE19 99% identical sequence in M. yongonense plasmid pMyong1 100% identical sequence in M. parascrofulaceum (plasmid?) mycP5 eccE5 eccA5 MCE locus on pMYCCH01 transposase M. chubuense plasmid pMYCCH01 5787 5786 5785 5784 5783 5782 5781 5780 5779 5778 5777 5776 80% 78% 60% 66% 63% 61% 64% 71% 52% 50% 50% 49% yrbE1A yrbE1B mce1A mce1B mce1C mce1D lprK mce1F M. tuberculosis Mce1 Rv0175 Rv0176 Rv0177 Rv0178 5775 mce1R fadD5 5788 transposase no more horizontal gene transfer! M. kansasii niche isolation? M. gastri M. ulcerans M. marinum M. canettii 70010 M. tuberculosis M. canettii 60008 M. xenopi cobF deletion Deletion of cobF (vitamin B12) in M. tuberculosis cobF M. canettii deletion in M. tuberculosis M. tuberculosis other methyltransferases may (partially?) compensate Gopinath et al. 2013. Future Microbiol 8:1405 The Great M. tuberculosis Schism pyruvate kinase SNP alanine dehydrogenase frameshift PhoR SNP cobL (+MK) deletion (RD9) more relaxed approach to host restriction? increasing species adaptation? M. tuberculosis may have evolved to rely on vitamin B12 provided by the host? niche adaptation • bioavailability of B12 in primates versus ruminants? • effect of diet – vegetarian versus meat-eating? • gut microbiome? The optional metabolome of vitamin B12 AMINO ACID BIOSYTHESIS MetH homocysteine methylmalonate (MutAB) MetE propionyl CoA methylcitrate (PrpCD) methionine DNA REPLICATION succinate deoxyribonucleotide NrdEF NrdZ ribonucleotide B12-independent B12-dependent ENERGY Lineage 5 Lineage 6 Lineage 4 22 independent SNPs and frameshifts predicted to impair function of MetH Lineage 2 Lineage 3 reduced reliance on B12-dependent pathways? Lineage 7 Lineage 1 post-Neolithic? human lung niche adaptation mycobacteria freely exchanging flexible functionality immunological vomiting niche isolation no turning back (no horizontal transfer) industrial remediation transmission cycle