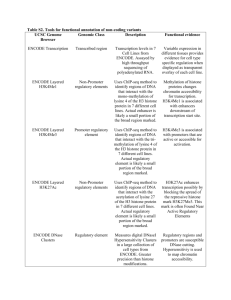
Presenting: On the immortality of television sets: *function* in the
advertisement

Presenting: On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE AKA: why the ENCODE project is full of it by Matthew Oberhardt What is ENCODE? •Attempt to find all functional elements of the human genome •huge international consortium, 10 years running •exome = 1.5% of human DNA •How much of the rest of it is garbage, vs. being useful ‘junk’ or fully functional? •pilot phase ended 2007 •production phase, 2007 – 2012 (with first major results published in 2012), and funded by $80 million in grants over 4 years •attempt to answer questions like: why are 88% of diseaseassociated SNPs in non-coding DNA regions? What did ENCODE do? mapped: RNA transcribed regions protein coding regions transcription factor binding sites chromatin structure DNA methylation sites performed assays on all of these biological areas in “tier 1,” “tier 2”, and “tier 3” cells – different standard cell types provide 1640 ‘datasets’ designed to annotate functional elements in the human genome ENCODE datatypes: Major findings: • 80.4% of the human genome participates in at least one biochemical RNA- and/or chromatin-associated event in at least one cell type (i.e., are ‘functional’ according to ENCODE) • Primate specific elements are in general negatively selected (fig 1) • classified chromatin states into groups with different promoter functionalities, and correlated RNA sequence production and processing to these chromatin states (showing that “most” variation in RNA expression can be explained by chromatin states). • found (or just repeated known information?) that most diseaserelated SNPs lie outside of coding regions But-- There are some problems with encode... On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE “Unless a genomic functionality is actively protected by selection, it will... cease to be functional. The absurd alternative, which unfortunately was adopted by ENCODE, is to assume that no deleterious mutations can ever occur in the regions they have deemed to be functional. Such an assumption is akin to claiming that a television set left on and unattended will still be in working condition after a million years because no natural events... can affect it.” On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE But let’s back up... On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE Major criticisms of ENCODE: (1) using the ‘causal role’ definition of biological function (2) committing the logical fallacy of ‘affirming the consequent’ (3) using analytical estimates that yield biased errors and inflate functionality estimates (4) favoring statistical sensitivity over specificity (5) emphasizing statistical significance rather than the magnitude of an effect Criticism 1: using the ‘causal role’ definition of biological function Two biological concepts of function: (1) The ‘causal role’ definition - a functional element is a genome segment producing a protein or an RNA or displaying a reproducible biochemical signature (e.g., protein binding) (2) The ‘selected effect’ definition – for a trait, T, to have a biological function F, it must (1) originate as a reproduction’ of some prior trait that performed F (or some similar function) in the past, and (2) T exists because of F. Example: a sequence similar to TATAAA can easily arise by chance, and will certainly bind transcription factors (being similar to the TATA box). It is therefore functional in the ‘causal role’ sense but not in the ‘selected effect’ sense. Similarly, the human heart has the ‘causal role’ of producing sounds, but its selected effect is pumping blood... Criticism 1: using the ‘causal role’ definition of biological function Bottom line: If a sequence doesn’t show signs of selection, it cannot be functional in the ‘selected effect’ manner, which is the only one that really counts. (this is a very strong statement...) Criticism 1: using the ‘causal role’ definition of biological function How, then, to detect selection? can have positive selection, purifying selection, or recently evolved speciesspecific elements. some of these can be subtle & hard to detect. SO – likely that more than 9% of the human genome is functional (what is currently thought) BUT – 80% is too high. Comparative genomics suggests that <15% of the genome is under evolutionary selection Therefore, % of functional elements should be below that... “ENCODE Incongruity”, that a biological function can be maintained without selection. Criticism 1: using the ‘causal role’ definition of biological function Why single out transcription as a function? You could also say ‘acted on by DNA polymerase’ is a function, in which case 100% of the genome is functional! ENCODE also uses this wrong definition of functionality wrongly... Criticism 2: committing the logical fallacy of ‘affirming the consequent’ The Fallacy: 1. if P then Q. 2. Q. 3. Therefore, P. Example: A random sequence binds a transcription factor; this does not necessarily result in transcription. However, the ‘binding’ property would be enough for ENCODE. In ENCODE, a DNA segment is ascribed ‘functionality’ if it is: (1) transcribed (2) associated with a modified histone (3) located in an open chromatin area (4) binds a transcription factor (5) contains a methylated CpG dinucleotide All of these are examples of affirming the consequent... Criticism 3: using analytical estimates that yield biased errors and inflate functionality estimates In ENCODE, a DNA segment is ascribed ‘functionality’ if it is: (1) transcribed (2) associated with a modified histone (3) located in an open chromatin area (4) binds a transcription factor (5) contains a methylated CpG dinucleotide All of these are examples of affirming the consequent... And continuing on this theme: Criticism 3: using analytical estimates that yield biased errors and inflate functionality estimates According to ENCODE, all of the below are (wrongly) considered functional: (1) 74.7% of genome that is transcribed – ALL OF WHICH IS CONSIDERED FUNCTIONAL • also, ENCODE used stem cells and cancer cells, both very transcriptionally active... • what about pseudogenes, introns, and mobile elements (non-functional)?? • Also, mapped RNA transcripts to DNA using a tool with 10% rejection rate (2) 56.1% that is associated with modified histones • A recent study showed 2% of histone modifications to affect function • ENCODE assigned functions to all histone modifications it analyzed (3) 15.2% that is found in open chromatin areas • ENCODE claims most open chromatin regions are functional transcription start sites • In fact, only 30% of open regions are even in the neighborhood of start sites (4) 8.5% that binds transcription factors • transcription sites are short, so many can occur by chance • better estimate is 0.28%, taking into account selection • Mean lengths of ENCODE ‘transcription factor binding sites’ are 824, 457, and 535 nucleotides, while most binding sitest are 6 – 14 bp!!!!! (5) 4.6% that is methylated CpG dinucleotides • ENCODE claims that 96% of CpG sites are methylated – not a sign of function, but merely that all CpG sites can be methylated! Evidence for purifying selection in ENCODE And the errors...: instead of using all SNPs, ENCODE used only the 1.3 million primate-specific ones of >=200bp ***By doing this, they removed everything that is of interest functionally!!! then, more processing left 82% of segments smaller than 100bp, with a median of 15bp, so: inferences in part using ~85,000 alignment blocks of 1bp and ~76,000 of 2bp... other problems with the controls... (they were longer, etc.) but in the end, the ENCODE-containing samples had a frequency 0.20% lower than control (hence negative selection!!). the pval was strong because there were so many datapoints (4e-37). IS THIS BIOLOGICALLY MEANINGFUL??? (stat test also probably didn’t take into account dependence of variables, and there are other possible causes of the 0.20% laid out) Evidence for purifying selection in ENCODE (CODING) allele frequency for primate-specific elements. this is the evidence for negative selection derived allele frequency Criticism 4: favoring statistical sensitivity over specificity (Just covered as well...) Criticism 5: emphasizing statistical significance rather than the magnitude of an effect Junk DNA ENCODE would have us think that “Junk DNA is Dead” A few distinctions: (1) Having a potential future function does NOT mean that a DNA segment is functional (hence ‘junk’, not ‘garbage’) (2) evolution will drive towards a mostly functional genome only if genome size is a significant negative selector & if the population size is huge – in humans neither are true (in bacteria they are), hence we expect a lot of junk. Big vs. Small science What is the function of ‘big science’? --to generate massive amounts of reliable & easily accessible data BUT – wisdom is best gained from small science... Take Home messages • selection is a *must* in ascribing a function to a gene. (is this strictly true?) • don’t affirm the consequent • don’t believe everything you read, even in prestigious journals... resistance is growing, as are multiply resistant strains reverse-incentive for drug companies to produce antibiotics, esp. narrow spectrum ones drugs today are very safe – high hurdle! penicillin wouldn’t have passed current standards! current Ab’s are off-patent & thus cheap, so doctors don’t want to use expensive new Ab’s infections present with vague symptoms usually... broad spectrum Ab’s are the best bet. Ab’s actually cure disease after a short run – not so good for $$ closing pipelines mean the intellectual base is scattering –we can’t just turn on the tap again!!