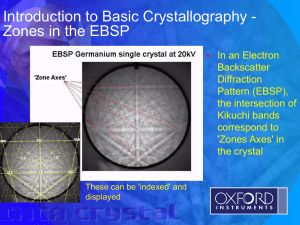
Fig. 1 - Open Access Crystallography
advertisement

Putting 3D-print files of crystallographic models into open access trevor.snyder@3dsystems.com International Advisory Board of the Crystallography Open Database & T. J. Snyder Modern crystallography has come a long way over the past 100 years; from a model of the very first crystal structure ever elucidated [1], Fig. 1, all the way to permanent URIs (on the Internet) for the structures of more than 250,000 small molecules and small to medium sized unit cell materials [2-4], Fig. 2. With the Crystallographic Information Framework (CIF,*.cif), there is a generally accepted and well documented “computer readable language of crystallography” [5]. Low cost 3D printing which utilizes the STereoLithography (STL,*.stl) format of 3D Systems Corporation became available recently, Fig. 3. It meets (and exceeds) the quality and low-cost maintenance needs of crystallographers (and the general public) for less than $1,000 [6]. Reduced cost and increased performance trends are expected to continue within the next few years, Fig. 4. Fig. 2 Promotional coffee mug from the International Advisory Board of the COD with the permanent URI of the caffeine molecule. We put crystallographic data into the public domain; it is up to you to use them and also to upload your own data to our website [2]. Fig. 1 Hard sphere model of the diamond structure by W. H. Bragg, Museum of the Royal Institution, London, photographed by Prof. André Authier. Taken from: http://blog.oup.com/2013/08/100th-anniversary-first-crystalstructure-determinations-bragg/#sthash.50BE8wiT.6mYKaBKB.dpuf and properly acknowledged below. Fig. 3 An inexpensive multi-color 3D printer from 3DSYSTEMS (less than $1,000), ref. [6], on the basis of their Plastic Jet 3D printing technology. Realizing that crystallographers now need conversion programs from CIF to STL to make good use of these recent developments, Werner Kaminsky incorporated such a converter into his well known WinXMorph program and created the CIF2VRML program from scratch [7]. While CIF2VRML reads small and macro-molecule *.cif files, displays them in the standard virtual reality format (VRML) and allows for exports into *.stl files, WinXMorph does the same (and much more) for crystal morphologies. Fig. 4 Mid 2012 “expectations versus time” graph of the Gartner group, showing 3D printing at the “peak of inflated expectations” and extrapolating to its “plateau of productivity” in 2017 (to 2022 at the latest). Figure 5 shows an annotated flow chart for the 3D printing of a model of a sugar molecule (of the kind that some of you may like to put into your coffee, i.e. sucrose). A partial flow chart for the 3D printing of a sucrose crystal complements this figure. We are committed to put all produced *.stl files into open access over time, beginning at a COD related project site that focuses on education [8]. Crystallography Open Database data_sugar_for_your_coffee solid sugar_for_your_coffee STL _chemical_formula_sum facet normal -0.13 -0.13 -0.98 outer loop vertex -16.25 14.23 -5.82 vertex -16.25 12.06 -5.54 vertex -18.42 14.23 -5.54 endloop endfacet facet normal 0.13 0.13 -0.98 outer loop vertex -16.25 14.23 -5.82 vertex -16.25 16.40 -5.54 vertex -14.08 14.23 -5.54 endloop endfacet… endsolid sugar_for_your_coffee STL 'C12 H22 O11' http://www.crystallography.net CIFs from open access databases, e.g. the COD (both figures to the right from ref. [4]) or its little “educational offsprings” (below, ref. [8]) [8a] [8b] loop_ _atom_site_type_symbol _atom_site_fract_x _atom_site_fract_y _atom_site_fract_z C 0.1310(2) 0.3413(3) 0.87541(17) C 0.2853(2) 0.3437(3) 0.99294(17) H 0.2730 0.4343 1.0421 C 0.4450(2) 0.3665(3) 0.93456(17) H 0.4824 0.2702 0.9045 C 0.3720(2) 0.4730(3) 0.82356(19) H 0.3889 0.5780 0.8527 C -0.0439(2) 0.4042(3) 0.89730(19) H -0.0222 0.4998 0.9419 H -0.1256 0.4222 0.8166 C 0.4528(2) 0.4517(3) 0.7107(2) H 0.3838 0.5085 0.6399 H 0.5717 0.4931 0.7306 C 0.2044(2) -0.0172(3) 0.64090(19) H 0.2815 -0.0248 0.7258 C 0.0583(2) -0.1374(3) 0.6261(2) H -0.0112 -0.1367 0.5386 C -0.0643(2) -0.1014(3) 0.71470(19) H 0.0024 -0.1084 0.8028 C -0.1356(2) 0.0589(3) 0.68727(18) H -0.2070 0.0621 0.6003 C 0.0147(2) 0.1767(3) 0.70012(17) H -0.0375 0.2754 0.6725 C 0.3159(2) -0.0370(3) 0.54289(19) H 0.4141 0.0335 0.5609 H 0.3642 -0.1386 0.5490 O 0.18374(14) 0.4395(2) 0.78776(13) O 0.10890(15) 0.18775(17) 0.82869(12) O 0.13181(16) 0.13534(19) 0.62240(13) O -0.12087(17) 0.2987(2) 0.97007(14) H -0.1435 0.2194 0.9306 O 0.29571(17) 0.2162(2) 1.07411(13) H 0.3169 0.1401 1.0370 O 0.58915(15) 0.4375(2) 1.02170(14) H 0.6818 0.3941 1.0197 O 0.46070(18) 0.2959(2) 0.67330(16) H 0.3607 0.2599 0.6566 O -0.24737(17) 0.0983(2) 0.77112(14) H -0.3239 0.1577 0.7355 O -0.2031(2) -0.2133(3) 0.69108(16) H -0.2344 -0.2307 0.7567 O 0.1446(2) -0.2792(2) 0.6518(2) H 0.0703 -0.3464 0.6462 O 0.2142(2) -0.0121(3) 0.41865(14) H 0.2172 0.0779 0.4012 … _space_group.point_group_H-M '2' more 3D print files downloadable in the future Fig. 5 Annotated flow charts for the 3D printing of models of a sucrose molecule (complete) and crystal (partial). loop_ _exptl_crystal_face_index_h _exptl_crystal_face_index_k _exptl_crystal_face_index_l _exptl_crystal_face_perp_dist -1 -1 0 0.51 -1 -1 1 0.584 -1 0 0 0.5681 … Werner’s downloadable software [7] (donations welcomed – otherwise free) ~ $5 currently, 1/10 of that in the near future Sure, you have much better uses for these kinds of models ! [1] W. H. Bragg and W. L. Bragg, The structure of diamond, Proc. R. Soc. Lond. A 89, 277 (1913) [2] www.crystallography.net; http://cod.ibt.lt/, http://cod.ensicaen.fr/, http://qiserver.ugr.es/cod/, http://nanocrystallography.org , and http://nanocrystallography.research.pdx.edu [3] S. Gražulis, D. Chateigner, R. T. Downs, A. F. T. Yokochi, M. Quirós, L. Lutterotti, E. Manakova, J. Butkus, P. Moeck, and A. Le Bail, Crystallography Open Database – an open-access collection of crystal structures, J. Appl. Cryst. 42, 726 (2009); http://journals.iucr.org/j/issues/2009/04/00/kk5039/kk5039.pdf, grazulis@ibt.lt [4] S. Gražulis, A. Daškevič, A. Merkys, D. Chateigner, L. Lutterotti, M. Quirós, N. R. Serebryanaya, P. Moeck, R. T. Downs, and A. Le Bail, Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration, Nucleic Acids Research 40, D420 (2012); http://nar.oxfordjournals.org/content/40/D1/D420.full.pdf+html, grazulis@ibt.lt [5] S. R. Hall, F. H. Allen, and I. D. Brown, The Crystallographic Information File (CIF): A New Standard Archive File for Crystallography, Acta Cryst. A 47, 655 (1991); http://www.iucr.org/iucr-top/cif/standard/cifstd1.html and http://www.iucr.org/resources/cif for continued updates as this standard progresses [6] January 6, 2014, press release: http://www.3dsystems.com/press-releases/3d-systems-recasts-consumer-3d-printing-experience-new-cuber-3 ; promotion: http://www.youtube.com/watch?feature=player_embedded&v=Osu5MC2PtMI ; http://www.3dsystems.com/press-releases/3d-systems-ups-prosumer-standards-new-sub-5000-cubeprotm-3d-printer ; http://www.3dsystems.com/press-releases/3d-systems-launches-cubifyr-20 [7] Prof. Werner Kaminsky: http://cad4.cpac.washington.edu/; CIF2VRML: http://cad4.cpac.washington.edu/cif2vrmlhome/cif2vrml.htm; WinXMorph: http://cad4.cpac.washington.edu/WinXMorphHome/WinXMorph.htm, kaminsky@chem.washington.edu [8] http://nanocrystallography.research.pdx.edu/search/edu/, pmoeck@pdx.edu; [8a] http://nanocrystallography.research.pdx.edu/media/sugar_molecule_stl.stl ; [8b] http://nanocrystallography.research.pdx.edu/media/sugar_morphology_stl.stl 3D printing, approx. $50 currently, 1/10 of that in the near future just kidding ? ? Support from NSF grant EEC-1242197 is gratefully acknowledged. Prof. André Authier is thanked for Fig. 1 which we took from his blog: http://blog.oup.com/2013/08/100thanniversary-first-crystal-structure-determinationsbragg/#sthash.50BE8wiT.6mYKaBKB.dpuf. No kidding, this work is one of our personal contributions to the UNESCO International Year of Crystallography (2014).