Lecture18 Biol302 Spring 2011
advertisement
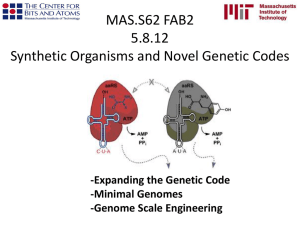
Polypeptide Chain Elongation http://www.molecularmovies.com/showcase/ An aminoacyl-tRNA binds to the A site of the ribosome. The growing polypeptide chain is transferred from the tRNA in the P site to the tRNA in the A site by the formation of a new peptide bond. The ribosome translocates along the mRNA to position the next codon in the A site. At the same time, – The nascent polypeptide-tRNA is translocated from the A site to the P site. – The uncharged tRNA is translocated from the P site to the E site. Elongation of Fibroin Polypeptides (A mRNA can have multiple Ribosomes Polypeptide Chain Termination Polypeptide chain termination occurs when a chain-termination codon (stop codon) enters the A site of the ribosome. The stop codons are UAA, UAG, and UGA. When a stop codon is encountered, a release factor binds to the A site. A water molecule is added to the carboxyl terminus of the nascent polypeptide, causing termination. No tRNA exists for stop codons! Dissociation upon finish of protein synthesis Fig1 The Genetic Code The genetic code is a nonoverlapping code, with each amino acid plus polypeptide initiation and termination specified by RNA codons composed of three nucleotides. Properties of the Genetic Code The genetic code is composed of nucleotide triplets. The genetic code is nonoverlapping. (?) The genetic code is comma-free. (?) The genetic code is degenerate. (yes) The genetic code is ordered. (5’ to 3’) The genetic code contains start and stop codons. (yes) The genetic code is nearly universal. YES :) A Triplet Code* A Single-Base Pair Insertion Alters the Reading Frame* A suppressor mutation restores the original reading frame.* Insertion of 3 base pairs does not change the reading frame.* Evidence of a Triplet Code: In Vitro Translation Studies Trinucleotides were sufficient to stimulate specific binding of aminoacyl-tRNAs to ribosomes. Chemically synthesized mRNAs containing repeated dinucleotide sequences directed the synthesis of copolymers with alternating amino acid sequences. mRNAs with repeating trinucleotide sequences directed the synthesis of a mixture of three homopolymers. Deciphering the Genetic Code You must know single letter codes and some triplets! What does Degree of Degeneracy Reflect? The Genetic Code Initiation and termination Codons – Initiation codon: AUG – Termination codons: UAA, UAG, UGA Degeneracy: partial and complete Ordered Nearly Universal (exceptions: mitochondria and some protozoa) Key Points Each of the 20 amino acids in proteins is specified by one or more nucleotide triplets in mRNA. (20 amino acids refers to what is attached to the tRNAs!) Of the 64 possible triplets, given the four bases in mRNA, 61 specify amino acids and 3 signal chain termination. (have no tRNAs!) Key Points The code is nonoverlapping, with each nucleotide part of a single codon, degenerate, with most amino acids specified by two to four codons, and ordered, with similar amino acids specified by related codons. The genetic code is nearly universal; with minor exceptions, the 64 triplets have the same meaning in all organisms. (this is funny) Do all cells/animals make the same Repertoire of tRNAs? The Wobble Hypothesis: Base-Pairing Involving the Third Base of the Codon is Less Stringent. Base-Pairing with Inosine at the Wobble Position Suppressor Mutations Some mutations in tRNA genes alter the anticodons and therefore the codons recognized by the mutant tRNAs. These mutations were initially detected as suppressor mutations that suppressed the effects of other mutations. Example: tRNA mutations that suppress amber mutations (UAG chain-termination mutations) in the coding sequence of genes. Making a (UAG) Mutation Translation of an amber (UAG) Mutation in the Absence of a Suppressor tRNA Translation of an amber Mutation in the Presence of a Suppressor tRNA Note it is amber su3…why????????? Translation of an amber Mutation in the Presence of a Suppressor tRNA If there was a single tRNATyr gene, then could one have a amber supressor of it? Historical Comparisons Comparison of the amino acid sequence of bacteriophage MS2 coat protein and the nucleotide sequence of the gene encoding the protein (Walter Fiers, 1972). Was this first???? Sickle-cell anemia: comparison of the sequence of the normal and sickle-cell alleles at the amino acid level and at the nucleotide level. Are the proteins produced a pure reflection of the mRNA sequence???? tRNA environment, protein modifications post-translationally Evolution? Alpha and Beta chain mutants…some of them Phylogenetic relationships How could we use GFP fluorescence to figure out-codon optimize GFP? CsCl centrifugation of DNA over time developed by Meselson and Stahl In class question (extra credit) for Quiz #4 Question 1: (0.5pts) Why does one add EtBr to CsCl gradients for the isolation of plasmid DNA? Question 2: (0.5pts-All or None credit) Is an 8kb supercoiled plasmid more dense than a 3kb supercoiled plasmid. Yes/No (circle one) Will an 8kb supercoiled plasmid have more EtBr bound to it? Yes/No (circle one) We will talk about this again in a later lecture: But CsCl gradients are not the same thing as Sucrose Gradients or Agarose Gel Electrophoresis. CsCl centrifugation of DNA over time N15 is heavier than N14-Can be resolved in CsCl pulse-chase Experiment: Incubator with N15 containing medium for time, then chase with N14 medium Expt 1 grows Slowly Expt 2 Bacteria Grow Faster Why? Why would they do 2 different growth rates? Experiment 1 Experiment 2 N14 N15 only N14 N15 only Fuse Results from Expt 1 and 2 Cell Divisions N14 N15 only Experiment 1 observations Watson-Crick Model N14 N15 only Does Expt 1 prove hybrid formation? N15 dsDNA N15 ssDNA Critical Experiment: Hybrid Strand Separation And CsCl centrifugation Looks like control below What about N14/N15 hybrid? N14 ssDNA N15 ssDNA Movie time To Know for Exam RNApol II TATAA CCATGG (Nco I site and Kozak Rule) ATG AGGT….splice GT……………A………polypyrimidine AG PolyA recog sequence AATAAA The Reasons why ATG is a single codon and TGG is a single codon. STRUCTURE AND FUNCTION OF ERYTHROPOIETIC TISSUE The RBCs ERYTHROPOIESIS (RBC PRODUCTION) Mature erythrocytes are derived from committed erythroid proginator cells through a series of mitotic divisions and maturation phases. Erythropoietin, a humoral agent produced mainly by the kidneys stimulates erythropoiesis by acting on committed stem cells to induce proliferation and differentiation of erythrocytes in the bone marrow. ERYTHROPOIESIS – Nucleated red cell precursors in the bone marrow are collectively called normoblasts or erythroblasts. – RBCs that have matured to the non-nucleated stage gain entry to the peripheral blood. – Once the cells have lost their nuclei, they are called erythrocytes. ERYTHROPOIESIS – Young erythrocytes that contain residual RNA are called reticulocytes. – Bone marrow normoblast proliferation and maturation occurs in an orderly and well defined sequence. • The process involves a gradual decrease in cell size, condensation and eventual expulsion of the nucleus, and an increase in hemoglobin production. BASIC BLOOD CELL MATURATION Nearly all hematopoietic cells mature in the manner shown below. For RBCs the nucleus is eventually extruded and the cytoplasm increase correlates with hemoglobin increase. ERYTHROPOIESIS GLOBIN SYNTHESIS ASSEMBLY OF HEMOGLOBIN Mutation: Changing Genetic Information ERYTHROPOIESIS • Reticulocytes are released from the bone marrow into the peripheral blood where they mature into erythrocytes , usually within 24 hours. • It is rare to see more than 1% reticulocytes in the peripheral smear from an adult, but common in healthy newborns. – They can be visualized more easily by staining with new methylene blue which allows for visualization of the remnants of the ribosomes on the endoplasmic reticulum. ERYTHROPOIESIS – Mature RBCs have a lifespan of 100-120 days and senescent RBCs are removed by the spleen. – 3 areas of RBC structure/metabolism are crucial for normal erythrocyte maturation, survival and function: • The RBC membrane • Hemoglobin structure and function • Cellular energetics