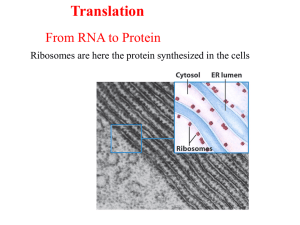
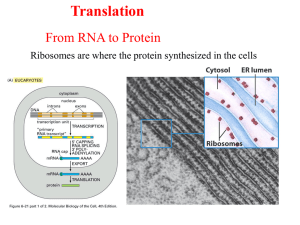
Chapter25_Outline

Chapter 25
Using the Genetic
Code
25.2 Related Codons Represent Chemically
Similar Amino Acids
• 61 of the 64 possible triplets encode 20 amino acids.
• Three codons ( stop codons ) do not represent amino acids and cause termination of translation.
Figure 25.01: All the triplet codons have meaning: 61 represent amino acids and 3 cause termination (stop codons).
25.2 Related Codons Represent Chemically
Similar Amino Acids
• The genetic code was frozen at an early stage of evolution and is nearly universal.
• Most amino acids are represented by more than one codon.
Figure 25.02: Some correlation of the frequency of amino acid use in proteins with the number of codons specifying the amino acid is observed.
25.2 Related Codons Represent Chemically
Similar Amino Acids
• The multiple codons for an amino acid are synonymous and usually related.
• third-base degeneracy – The lesser effect on codon meaning of the nucleotide present in the third (3′) codon position.
• Chemically similar amino acids often have related codons, minimizing the effects of mutation.
25.3 Codon –Anticodon Recognition
Involves Wobbling
• Multiple codons that represent the same amino acid most often differ at the third base position (the wobble hypothesis ).
Figure 25.03: Third bases have the least influence on codon meanings.
25.3 Codon –Anticodon Recognition
Involves Wobbling
• The pairing between the first base of the anticodon and the third base of the codon can vary from standard Watson-Crick base pairing according to specific wobble rules.
Figure 25.04: Wobble in base pairing allows G-U pairs to form between the third base of the codon and the first base of the anticodon.
25.3 Codon –Anticodon Recognition
Involves Wobbling
Figure 25.05: Codon–anticodon pairing involves wobbling at the third position.
25.4 tRNAs are Processed from Longer
Precursors
• A mature tRNA is generated by processing a precursor.
• The 5′ end is generated by cleavage by the endonuclease RNAase P.
• The 3′ end is generated by multiple endonucleolytic and exonucleolytic cleavages, followed by addition of the common terminal trinucleotide CCA.
Figure 25.06: The tRNA 3 end is generated by cutting and trimming reactions, followed by addition of CCA when this sequence is not coded.
25.5 tRNA Contains Modified Bases
• 81 examples of modified bases in tRNAs have been reported.
• Modification usually involves direct alteration of the primary bases in tRNA, but there are some exceptions in which a base is removed and replaced by another base.
Figure 25.07: Each of the four bases in tRNA can be modified.
25.5 tRNA Contains Modified Bases
• Known functions of modified bases are to confer increased stability to tRNAs and to modulate their recognition by proteins and other RNAs in the translational apparatus.
25.6 Modified Bases Affect Anticodon –
Codon Pairing
• Modifications in the anticodon affect the pattern of wobble pairing and therefore are important in determining tRNA specificity.
Figure 25.08: Inosine can pair with U, C, or A.
Figure 25.09: Modification to 2thiouridine restricts pairing to A alone because only one H-bond can form with G.
25.7 There Are Sporadic Alterations of the
Universal Code
• Changes in the universal genetic code have occurred in some species.
• These changes are more common in mitochondrial genomes, where a phylogenetic tree can be constructed for the changes.
Figure 25.11: Changes in the genetic code in mitochondria can be traced in phylogeny.
25.7 There Are Sporadic Alterations of the
Universal Code
• In nuclear genomes, the changes usually affect only termination codons.
Figure 25.10: Changes in the genetic code in bacterial or eukaryotic nuclear genomes usually assign amino acids to stop codons or change a codon.
25.8 Novel Amino Acids Can Be Inserted at
Certain Stop Codons
• The insertion of selenocysteine at some UGA codons requires the action of an unusual tRNA in combination with several proteins.
• The unusual amino acid pyrrolysine can be inserted at certain UAG codons.
• The UGA codon specifies both selenocysteine and cysteine in the ciliate Euplotes crassus .
Figure 25.12: SelB is an elongation factor that specifically binds tRNASec to a UGA codon that is followed by a stem-loop structure in mRNA.
25.9 tRNAs Are Charged with Amino Acids by Aminoacyl-tRNA Synthetases
• Aminoacyl-tRNA synthetases are a family of enzymes that attach amino acid to tRNA, generating aminoacyltRNA in a two-step reaction that uses energy from ATP.
• Each tRNA synthetase aminoacylates all the tRNAs in an isoaccepting (or cognate ) group, representing a particular amino acid.
25.9 tRNAs Are Charged with Amino Acids by Aminoacyl-tRNA Synthetases
• Recognition of tRNA by tRNA synthetases is based on a particular set of nucleotides, the tRNA
“ identity set, ” that often are concentrated in the acceptor stem and anticodon loop regions of the molecule.
Figure 25.13: An aminoacyl-tRNA synthetase charges tRNA with an amino acid.
25.10 Aminoacyl-tRNA Synthetases Fall into Two Classes
• Aminoacyl-tRNA synthetases are divided into class I and class II families based on mutually exclusive sets of sequence motifs and structural domains.
Figure 25.14: Separation of tRNA synthetases into two classes possessing mutually exclusive sets of sequence motifs and active-site structural domains.
Figure 25.16: Crystal structures show that class I and class II aminoacyl-tRNA synthetases bind the opposite faces of their tRNA substrates.
25.11 Synthetases Use Proofreading to
Improve Accuracy
• Specificity of amino acid-tRNA pairing is controlled by proofreading reactions that hydrolyze incorrectly formed aminoacyl adenylates and aminoacyl-tRNAs.
• kinetic proofreading – A proofreading mechanism that depends on incorrect events proceeding more slowly than correct events, so that incorrect events are reversed before a subunit is added to a polymeric chain.
25.11 Synthetases Use Proofreading to
Improve Accuracy
Figure 25.17: Aminoacylation of cognate tRNAs by synthetase.
25.11 Synthetases Use Proofreading to
Improve Accuracy
• chemical proofreading –
A proofreading mechanism in which the correction event occurs after the addition of an incorrect subunit to a polymeric chain, by means of reversing the addition reaction.
Figure 25.18: Proofreading by aminoacyl-tRNA synthetases.
25.12 Suppressor tRNAs Have Mutated
Anticodons That Read New Codons
• A suppressor tRNA typically has a mutation in the anticodon that changes the codons which it recognizes.
25.12 Suppressor tRNAs Have Mutated
Anticodons That Read New Codons
• When the new anticodon corresponds to a termination codon, an amino acid is inserted and the polypeptide chain is extended beyond the termination codon.
– This results in nonsense suppression at a site of nonsense mutation, or in readthrough at a natural termination codon.
25.12 Suppressor tRNAs Have Mutated
Anticodons That Read New Codons
Figure 25.20: Nonsense mutations can be suppressed by a tRNA with a mutant anticodon.
25.12 Suppressor tRNAs Have Mutated
Anticodons That Read New Codons
• Missense suppression occurs when the tRNA recognizes a different codon from usual, so that one amino acid is substituted for another.
Figure 25.21: Missense suppression occurs when the anticodon of tRNA is mutated so that it responds to the wrong codon.
25.13 There Are Nonsense Suppressors for
Each Termination Codon
• Each type of nonsense codon is suppressed by tRNAs with mutated anticodons.
• Some rare suppressor tRNAs have mutations in other parts of the molecule.
Figure 25.22: Nonsense suppressor tRNAs are generated by mutations in the anticodon.
25.14 Suppressors May Compete with Wild-
Type Reading of the Code
• Suppressor tRNAs compete with wild-type tRNAs that have the same anticodon to read the corresponding codon(s).
• Efficient suppression is deleterious because it results in readthrough past normal termination codons.
• The UGA codon is leaky and is misread by Trp-tRNA at
1% to 3% frequency.
25.14 Suppressors May Compete with Wild-
Type Reading of the Code
Figure 25.23: Nonsense suppressors also read through natural termination codons, synthesizing polypeptides that are longer than the wild type.
25.15 The Ribosome Influences the
Accuracy of Translation
• The structure of the 16S rRNA at the P and A sites of the ribosome influences the accuracy of translation.
Figure 25.24: Any aminoacyl-tRNA can be placed in the A site, but only one that pairs with the anticodon can make stabilizing contacts with rRNA.
25.16 Frameshifting Occurs at Slippery
Sequences
• The reading frame may be influenced by the sequence of mRNA and the ribosomal environment.
• recoding – Events that occur when the meaning of a codon or series of codons is changed from that predicted by the genetic code.
– It may involve altered interactions between aminoacyl-tRNA and mRNA that are influenced by the ribosome.
25.16 Frameshifting Occurs at Slippery
Sequences
• Slippery sequences allow a tRNA to shift by one base after it has paired with its anticodon, thereby changing the reading frame.
• Translation of some genes depends upon the regular occurrence of programmed frameshifting .
Figure 25.26: A 11 frameshift is required for expression of the tyb gene of the yeast Ty element.
25.16 Frameshifting Occurs at Slippery
Sequences
Figure 25.25: A tRNA that slips one base in pairing with a codon causes a frameshift that can suppress termination. The efficiency is usually ~5%.
25.17 Other Recoding Events: Translational
Bypassing and the tmRNA Mechanism to
Free Stalled Ribosomes
• Bypassing involves the capacity of the ribosome to stop translation, release from mRNA, and resume translation some 50 nucleotides downstream.
Figure 25.28: In bypass mode, a ribosome with its P site occupied can stop translation.
Figure 25.27: Bypassing
25.17 Other Recoding Events: Translational
Bypassing and the tmRNA Mechanism to
Free Stalled Ribosomes
• Ribosomes that are stalled on mRNA after partial synthesis of a protein may be freed by the action of tmRNA , a unique RNA that incorporates features of both tRNA and mRNA.