Rhodococcus opacus - Engineering Student Services and Academic
advertisement

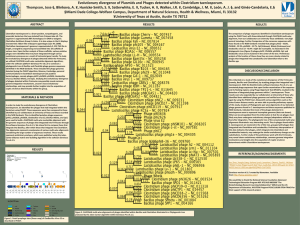
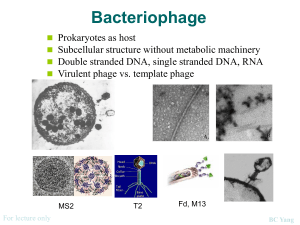
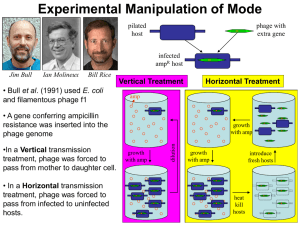
Rhodococcus opacus What is the Research Question? Armando Vital Rivera High School Brownsville ISD Dr. Kung-Hui (Bella) Chu Assistant Professor, Department of Civil Engineering (Environmental Engineering) Texas A&M University TYPE OF ENGINEERING Civil Engineering Lab Environmental research is conducted in the CVLB lab. Working with two of Dr. Chu’s PhD students, Myung Hee Kim and Do Gyun Lee Center for Phage Technology (CPT) lab Biology research is conducted in the Center for Phage Technology Working with Dr. Jason Gill and Dr. Ry Young BACKGROUND OF RESEARCH PROJECT Dr. Kung-Hui Chu Work Biodegradation and bioremediation of priority pollutants and emerging contaminants Molecular quantification of microbial risk in water Optimization of bioenergy production Application of bioretention for stormwater runoff management Advancing knowledge on microbial ecology of nitrogen and carbon cycles RELEVANCE OF THE RESEARCH Research Question A tale of two phages: Is phage DNA sequence highly conserved over time and space? Significance: Phage Genome Evolution THE LAB WORK Techniques used in molecular biology for working with the phages. Applied microbiology, virology, and environmental engineering. RESEARCH OBJECTIVE Isolate and characterize phages that infect Rhodococcus opacus. Collect activated sludge and soil samples that may contain phage. EXPLORATION What do we know about these phages? A phage, Ropa 4, was isolated in Germany over 20 years ago Three R. opacus phages were recently isolated in Chu’s laboratory The genomes of all four phages were sequenced at the CPT and found to be almost identical SURPRISE DISCOVERY! The DNA of the newly isolated phage is almost 100% identical to that of the phage isolated from Germany. Signifies a new finding in “Phage Genome Evolution” Approach: E3 teachers will repeat our work at other laboratory in order to rule out any cross contamination during isolation phages in Dr. Chu’s lab. BACKGROUND ON PHAGES Bacteriophages are viruses that infect bacterial cells. Phages cannot reproduce on their own. The phage hijacks the cell’s machinery to reproduce progeny. Phages are specific for their host bacteria. YouTube - T4 Virus infecting a bacteria. Rhodococcus opacus Rhodococcus opacus is a specific bacteria Belongs to the family Actinomycetes, related to Mycobacterium R. opacus is a rod, nonmotile, mycobacterium This bacteria was used as the host to isolate phage in Chu’s laboratory Impact of the Research Potentially open a new research direction in phage genomics. Some Rhodococcus opacus species are pathogens. A better understanding of phages specific to R. opacus can enhance the development of phage therapy. Some R. opacus are foaming bacteria in biological wastewater treatment processes. Phage treatment might be possible to minimize common sludge bulking problem. THE CHALLENGE FOR SCIENCE The cell wall of the bacteria is difficult disrupt. First step is to see which phages can actually infect R. opacus Rhodococcus-specific phages can lyse the cell. LYSIS PROCESS Lysis via a 3 component system in Mycobacteria A holin protein opens a pore in the cytoplasmic membrane The creation of the pore triggers the release of endolysin An esterase enzyme is also released that degrades the outer mycolic acid cell wall layer Prepare 4 litters of R2A broth (media/nutrients) Research Activities Phage Titration Prepare broth with 1.5% Agar (solid) Prepare broth with 0.8% Agar (gel) Collect soil samples Prepare petri dishes Add R. opacus to molten agar Collect phage from soil Pour bacterial lawn to petri dishes to achieve an even growth of bacteria Apply phage spots to bacterial lawn Proceed to full-plate titration of phage. RESEARCH ACTIVITIES Learn laboratory aseptic techniques when working with BL1 agents. Use ethanol as sanitizer Use flame to allow clean air to move upwards preventing bacteria from falling on the bench. Prepare soft agar overlay plates Prepare media of R2A with 1.5% Agar concentration and 0.8% agar concentration. Spot Titration of phage Pour a bacterial lawn Label bottom surface of plate with phage and lawn strain. In aseptic environment, transfer host culture into molten agar and vortex. Pour contents of tube onto agar surface. Allow agar to solidify. Spot Titration of Phage Apply phage spots to lawn. Aspirate phage dilution and deposit over corresponding label marked on plate. Allow plate to dry near a flame for 15 min Incubate plates, inverted, at optimal growth temperature until plaques appear in the lawn. Individual plaques may be counted to estimate the concentration of phage. EXPERIMENTS Collecting soil samples Preparing media (broth/plates) Enriching samples for phage Growing liquid cultures of bacteria Plating out phage Collecting plaques of phage on petri dishes DATA TO BE GENERATED Data that will help characterize the phages from the soil samples collected from a sewer facility and close to a gasoline station. The model used will be comparing the characteristics of newly found phages to the phage currently available. Future Implications Is the high similarity of phage sequences unique to Rhodococcus species? Does the high similarity of phage sequences present concern G+ pathogens? What is the implications of high conservation of phage sequences? SUMMARY Learn how to grow R. opacus bacteria. Collect soil samples from a sewer facility and close to a gasoline station. Isolate, and characterize newly found phages. Compare newly found phages to the phage found in Germany. CLASSROOM APPLICATION Combined lesson plan with Biology and Algebra 1 classes. Analyze and evaluate phage growth rate using functions and scatter plots. ACKNOWLEDGMENTS TAMU E3 Program National Science Foundation Nuclear Power Institute Texas Workforce Commission Dr. Kung-Hui (Bella) Chu The Center for Phage Technology Dr. Jason Gill and Dr. Ry Young Myung Hee Kim and Do Gyun Lee Andy Hernandez (partner) THANK YOU!