Review Problems #1 – 09/26/2012 For λ phage to infect a bacterium
advertisement

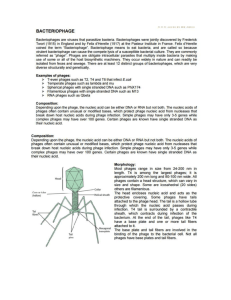
Review Problems #1 – 09/26/2012 1. For λ phage to infect a bacterium, it must first interact with receptors on the surface of the bacterium. Why do bacteria express receptors that allow λ to infect them? The proteins obviously have another purpose, and lacking these receptors is more reproductively limiting than allowing infection by λ. What types of receptors do you think are targeted by λ? On the surface of many bacteria Allow transport into cell Actually use Porin. Large beta-barrel protein through which molecules can diffuse. Present in the outer membrane of most Gram-negative and some Gram-positive bacteria, it is widely useful. Produced under the maltose operon – a key metabolic pathway, not some obscure receptor that bacteria could whimsically turn off. 2. Your current lab project is to PCR amplify your favorite gene and insert it into your expression vector. Unfortunately, you were hungover the day after Granlibakken, and you left out your freezer box on the bench. Now, your entire collection of restriction enzymes and ligases is compromised and you are getting impossibly low transformation efficiency into your bacteria. Rather than admit your expensive mistake to your boss, and being a keen molecular biologist, you start thinking of alternatives. It dawns on you that DNA recombination is significantly more efficient than DNA digestion and ligation. As you are particularly well-versed in λ phage lysogeny, you notice a few similarities: Your PCR product is a linear piece of DNA, kind of like the packaged λ genome Your expression vector is a circular plasmid, kind of like the bacterial chromosome λ integration occurs in a very site-specific manner, kind of like the specificity achieved with different restriction enzymes What elements from λ could you incorporate your cloning scheme to catalyze the integration of your PCR product into your expression vector? (After you have figured this out, give yourself a pat on the back and rush to the patent office. You have invented Invitrogen’s Gateway cloning system.) attP sites flanking PCR product Add the ‘cos’ site to circularize attB site in vector Introduce λ integrase to catalyze recombination 3. You are interested in host factors that promote lysogen formation. How would you go about isolating mutants? Mutagenize bacteria, isolate clones, infect with phage and look for clear plaques What types of genes would you find? Would they all directly interact with lambda’s fate decision machinery? You would pull out cap, any metabolic genes or transporters that normally lead to HIGH glucose, and constitutively active recA, so no they won’t all be directly involved. Mutations in the attB site Will you obtain mutants in all the positive regulators of lysogeny? Probably not. Adenyl cyclase and key metabolic genes are required for viability Your PI informs you that for his next grant he wants to investigate the SOS response. How would you please the PI and ensure your continued funding by distinguish between the SOS and metabolic mutants you have isolated in your screen? You could infect bacteria growing on different carbon sources to weed out some of the metabolic genes that may be having a context dependent effect on glucose levels. Transfect the bacteria with a plasmid expressing a dominant negative recA to identify the mutants with a constitutive SOS response. There is a newly discovered phage - alpha. Because you are a mad scientist, secretly bent on destroying the research of your competitor, you want to engineer a phage that will secretly infect all of the cultures of your competitor lab so that every time they try to grow a culture of bacteria, their bacterial cultures are lysed. Since this is a new phage, its genes weren’t discovered in the 1970s, it’s your job to identify the genes in alpha. You have already performed a screen to identify essential genes in alpha, and have many mutants, but now you need to know how many different genes you’ve identified. 1) What types of test can you perform to determine this? Complementation test 2) Should this test be performed at high or low MOI? Why? Should be high MOI to ensure that each bacteria has two phages. 3) You have a tube of phage. You have done a plaque assay with 10 serial dilutions. The results are listed in the diagram below. You know that you will be doing your experiment with 10^8 bacteria. If you want to infect your bacterial cultures at an MOI of 5, what volume of your phage stock would you need to add to your flask of bacteria? This problem is setup so that your concentration of phage is 3 x 10^10 phages/mL so that when you do serial dilutions you get 300, 30, 3 phages on your plates. MOI 5 for 10^8 bacteria = 5 x 10^8 phages. 5 X 10^8/3x 10^10 phages/mL = .0166 mL = 16.7 uL. You want to add 16.7 uL of phage to get MOI of 5. 4) Now you know what volume of phage to add, you proceed with the test and get the following results for your complementation test. 1 1 2 3 4 5 6 7 - + + + - - - - + - + - + - + + - - - + - + - - - - - 2 3 4 5 6 7 - 2) List all the complementation groups Group A: 1, 7, 5 Group B: 2, 4 Group C: 3, 7 Group D: 6 3) What’s up with mutant number 6? Either it’s a dominant negative mutation, or a gain of function mutation. 4)What would you do to validate your hypothesis? Do a co-infection with wildtype to be to make sure that WT can complement it. If it is dominant negative co-infection with wild-type will not rescue the mutant phenotype of #6. 4) Mutant 7 does not complement mutant 1 or 3, while they complement each other. Can you give an explanation for why this would happen? Possible answers: 1) There are mutations in two separate genes. 2) There could be mutations in two separate domains of the same protein. If 3 is required for function Y, and 1 is required for function X, and mutation 7 is in a domain required for X and Y, this would also explain your results. A real life example is disheveled. There are mutations in disheveled that either disrupt canonical wnt signaling, or PCP or both. A PCP and a signaling mutant allele will complement eachother, even though they are in the same gene. 4B) Can you think of another explanation for why 7 would fail to complement another gene? Two genes in the same pathway (ie, A -> B -> C), or mutations in regions of the genome that don’t act in trans (ie, promoters or operators) You could make a branched pathway with 1 and 3 in parallel pathways, but both lead to 7 later on and the non-complementation is due to reduced gene dosage down either pathway. 5) How would you test your hypothesis? Complementation analysis probably wouldn’t work because it would be hard to interpret the results from triple infection. A better strategy would be to try to isolate the mutation by back-crossing, and then re-do the complementation analysis. You could also map mutation 1 and 3 to see if they map to the same location. Additional Information and further reading: 1) http://en.wikipedia.org/wiki/Lamda_phage I know what you’re thinking…. Wikipedia, really genetics TAs? But, the Wikipedia page is actually fairly comprehensive and has a nice list of all of the different phage genes, and what they do. 2) Dodd, J.B. Shearwin; Egan, J.B.; Egan, JB (2005). "Revisited gene regulation in phage lambda". Curr Opin Genet Dev 15 (2): 145–152. doi:10.1016/j.gde.2005.02.001 Provides a nice overview of phage interactions 3) Friedman, D.I.; Court, D.L. (2001). "Bacteriophage lambda; alive and well and still doing its thing". Current Opinion in Microbiology 4 (2): 201–207. Brief overview of phages in the modern area of biology, because you know, not all things regarding phages were discovered before you were born. 4) K. Baek, S. Svenningsen, H. Eisen, K. Sneppen, S. Brown Single-cell analysis of lambda immunity regulation J Mol Biol, 334 (2003), pp. 363–372 This study demonstrated that the vast majority of phages released from λ lysogens in the absence of the SOS system are mutants defective in cI activity or expression. The authors used a gfp insertion downstream of cI to confirm OL involvement in cI autoregulation and found a large amount of natural variation in cI expression among lysogens. (see there are still interesting things regarding phages happening in relatively current science!!) 5) I.B. Dodd, K.E. Shearwin, A.J. Perkins, T. Burr, A. Hochschild, J.B. EganCooperativity in longrange gene regulation by the lambda cI repressor Genes Dev, 18 (2004), pp. 344–354 In this study, repression of PRM (but not PR) by physiological cI levels was eliminated by mutation of OL3. The authors demonstrated interaction between cI CTDs at OL3 and OR3 with the use of a P22:λ hybrid repressor. Modelling was used to estimate long-range cooperativities and to predict major cI–DNA species. Looping efficiency suggests that the entropic costs of long-range DNA looping in vivo might be 80-fold less than those in vitro. (More on the mechanism of cI repression)