Phage Discovery Lab
advertisement

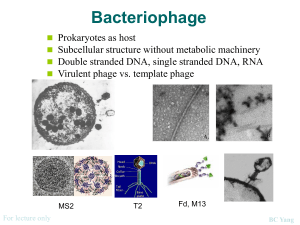
Expansion of Gonzaga University’s phage research program Marianne Poxleitner, Kirk Anders, Amanda Braley, and Ann-Scott Ettinger Department of Biology, Gonzaga University, Spokane, WA 99258 The Challenge Phage Discovery Lab Genetics Lab Fall Family Phage Symposium Provide all introductory biology students at Gonzaga with a lab course that provides a research experience The In Situ portion of the phage course was taught to ~360 incoming science freshmen as the introductory biology lab course serving many majors: The In Silico portion was taught as two modules within a Genetics Laboratory course (BIOL 207L). This year, ~90 students participated. Select students presented their research during a mini phage symposium scheduled during the annual “Fall Family Weekend” activities. After observing the student enthusiasm and outcomes of the first two years of the SEA PHAGES course, our colleagues asked if we could modify the course to serve as the lab for our large introductory biology course in our new curriculum. But how to do it in one day per week? What about genome annotation? Can we keep the “magic” at an industrial scale? Our Strategy • • • • • • • • • Biology Biochemistry Human Physiology Nursing Premed (non-majors) Computer Science (a few) Implementation We converted the In Situ and In Silico portions of a SEA PHAGES course into a progression of experiences that are vertically integrated in our curriculum. Figure 3. Lots of enrichment cultures shaking at room temperature. Table 1. Semester lab activity schedule by week. Week 1 The Phage Discovery Lab serves a large number of freshmen students, both science and health science majors. 2 Plate enrichment supernatant in dilution series 3 7 Achieve these activities: Test putative plaques Plaque-purify at least three rounds Determine titers of plaque lysates Flood “web” plate to produce medium titer lysate Titer medium titer lysate (a few exceptional students prepare high titer lysate) 8 Mount samples for transmission electron microscopy 9 DNA prep 4 The Genome Annotation and Phage DNA Cloning modules in the 200-level Genetics Laboratory course serve all biology and biochemistry majors. 5 6 The Advanced Phage Research lab serves a small number of interested 3rd and 4th year students. In addition to extra supplies from the SEA, we obtained funding from the NSF-TUES program and from Gonzaga University to supplement lab fees and equip laboratory classrooms to teach these courses. Phage Research Lab Activity Collect soil, inoculate enrichment culture 90 students/year Phage Discovery Lab 360 introductory students/year Figure 1. Gonzaga University’s three tiered phage research program. • One freshman from the Phage Discovery Lab presented background materials and details about isolation, purification, and analysis of her phage Figure 6. Genetics students showing attitude. Genome Annotation Module (5 weeks) • Finding genes in DNA: 6-frame translations, ORFs, RBS, coding potential • Learn button-pushing in DNA Master. Annotate 1-2 genes as a class • Annotation in teams of two pairs. Each team annotates 6-10 genes • Class presentations of gene calls and functions • Teams work on exploratory projects (find other elements/repeats, compare sequences and phylogenies, propose own question, etc.) A • A previous SEA PHAGES student presented bioinformatics results from the previous year on the sequenced phage Phoxy • Six freshmen presented Phage Discovery posters assembled the previous semester during the Fall Family Weekend poster session (usually reserved for upperclassmen presenting faculty research) • 65 attendees were able to see the cohesive nature of our phage program. Students got to see what phage fun was was to come and parents observed students presenting work done as part of our Biology curriculum B 10 DNA prep 11 Restriction digest 12 Agarose gel electrophoresis 13 Restriction fragment gel analysis • MTL DNA prep again, Sal I digest, ligate to pBluescript 14 Student poster presentations • Transform competent E. coli, plate to Amp + X-gal plates 15 Final exam and assessment • Isolate plasmids from transformants, measure insert size Phage DNA Cloning and Sequencing Module (5 weeks) • Students recover their own phage from archived lysate • Send clone for Sanger sequencing with T3 and T7 primers • • • • • • • 12 lab sections fall semester, and 12 spring semester Meet one day a week for 3 hours 16 students per lab work in pairs to isolate phage Taught by faculty Hire teaching assistants from previous semesters Utilized lab technique videos and online quizzes to save class time Incubate cultures at room temp to slow growth and facilitate large numbers of plates • Purification shortcuts include streak plates outside of class time • Use MTL for TEM and DNA isolation Assessment • SEA CURE -- compare to full-year courses • Pre/post concept test • Open-ended survey to probe scientific thinking, curiosity, and enthusiasm (we are studying these for patterns, hints, and ideas about outcomes we might try to measure in the future) • Analyze sequence, assign tentative cluster, propose which phage genomes should be sequenced in future Advanced Phage Research Lab To begin Fall 2014. Students who have completed Genetics Lab may apply for this course, and take it multiple times if they wish. Purpose • want to stay involved in phage research -or- • want research experience in hopes of eventually getting an undergraduate research position • Run like a research group. Research outside of class time is required. • A mixture of faculty-organized and student-generated projects to choose from • Molecular and/or bioinformatics methods to be used • Work in small teams or individually • Students write proposals, design experimental approaches, present during informal lab meetings, formal presentations at end Figure 2. Students participating in Phage Discovery Lab. Figure 5. Fall 2012 shipment. That’s a lot of boxes! Figure 9. Finding plaques during Faculty Phage training. Faculty Phage Training Enhance faculty understanding of the Phage Discovery Lab goals • Offered summer training to faculty at local universities and community colleges to share materials and strengthen the research oriented goals of the course, and discourage teaching it like a “cookie- cutter” lab. • Assembled a “Faculty Phage” training manual to highlight the research goals of each period and suggest ways to facilitate discussions and avoid lectures. • Additional training with faculty teaching freshman Phage Discovery labs at Gonzaga will focus on reducing lecturing and encourage using a “lab meeting” type approach in the classroom. • Reinforce these goals with weekly meetings to make sure everyone is on task and teaching in a manner that is in keeping with the HHMI vision of the SEA PHAGES course. To provide opportunity for students to participate in independent-style research. These are students who: Tentative plan Figure 4. Students doing a restriction enzyme digest activity • A senior presented cluster typing and sequencing results for the Genetics Lab Figure 7. Blue/white screening (A) and Sanger sequences (B) from molecular biology module. 10-20 students/yr Phage Genomics and Molecular Biology Modules Biology majors Biochemistry Premed (non-majors) Acknowledgments Lu Barker, Kevin Bradley (SEA HHMI) and Debbie Jacobs-Sera, Welkin Pope, Dan Russell, Graham Hatfull (Pittsburgh Bacteriophage Institute) for training, assistance, and encouragement David Asai, Cheryl Bailey, Lu Barker, and HHMI for supplies and other encouragement and support NSF TUES program for equipment and support Gonzaga University for support and enthusiasm Leslie Jaworski and David Lopatto for CURE survey results