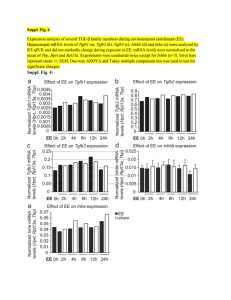
Basic Principles of RT-qPCR | Thermo Fisher Scientific - IN 1 of 4 https://www.thermofisher.com/in/en/home/brands/thermo-scientific/mo... Home › Brands › Thermo Scientific › Molecular Biology › Molecular Biology Resource Library › Molecular Biology Resource Library › Spotlight Articles–Thermo Scientific Molecular Biology › Basic Principles of RT-qPCR Basic Principles of RT-qPCR Introduction to RT-qPCR RNA as the Starting Material Quantitative reverse transcription PCR (RT-qPCR) is used when the starting material is RNA. In this method, RNA is first transcribed into complementary DNA (cDNA) by reverse transcriptase from total RNA or messenger RNA (mRNA). The cDNA is then used as the template for the qPCR reaction. RTqPCR is used in a variety of applications including gene expression analysis, RNAi validation, microarray validation, pathogen detection, genetic testing, and disease research. One-step vs. Two-step RT-qPCR RT-qPCR can be performed in a one-step or a two-step assay (Figure 1, Table 1). One-step assays combine reverse transcription and PCR in a single tube and buffer, using a reverse transcriptase along with a DNA polymerase. One-step RT-qPCR only utilizes sequence-specific primers. In two-step assays, the reverse transcription and PCR steps are performed in separate tubes, with different optimized buffers, reaction conditions, and priming strategies. Click image to enlarge Figure 1. One-Step vs. Two-Step RT-qPCR. Table 1. Advantages and Disadvantages when using one-step versus two-step assays in RT-qPCR Advantages One-step Two-step Disadvantages Less experimental variation since both reactions take Impossible to optimize the two reactions separately place in the same tube Less sensitive than two-step because the reaction Fewer pipetting steps reduces risk of contamination conditions are a compromise between the two combined Suitable for high throughput amplification/screening reactions Fast and highly reproducible Detection of fewer targets per sample A stable cDNA pool is generated that can be stored for The use of several tubes and pipetting steps exposes long periods of time and used for multiple reactions the reaction to a greater risk of DNA contamination The target and reference genes can be amplified from the Time consuming same cDNA pool without multiplexing Requires more optimization than one-step Optimized reaction buffers and reaction conditions can be used for each individual reaction Flexible priming options 8/31/2019, 3:29 PM Basic Principles of RT-qPCR | Thermo Fisher Scientific - IN 2 of 4 https://www.thermofisher.com/in/en/home/brands/thermo-scientific/mo... Reverse Transcription in RT-qPCR Choosing total RNA vs. mRNA When designing a RT-qPCR assay it is important to decide whether to use total RNA or purified mRNA as the template for reverse transcription. mRNA may provide slightly more sensitivity, but total RNA is often used because it has important advantages over mRNA as a starting material. First, fewer purification steps are required, which ensures a more quantitative recovery of the template and a better ability to normalize the results to the starting number of cells. Second, by avoiding any mRNA enrichment steps, one avoids the possibility of skewed results due to different recovery yields for different mRNAs. Taken together, total RNA is more suitable to use in most cases since relative quantification of the targets is more important for most applications than the absolute sensitivity of detection1. Primers for Reverse Transcription Three different approaches can be used for priming cDNA reactions in two-step assays: oligo(dT) primers, random primers, or sequence specific primers (Figure 2, Table 2). Often, a mixture of oligo(dT)s and random primers is used. These primers anneal to the template mRNA strand and provide reverse transcriptase enzymes a starting point for synthesis. Figure 2. Four different priming methods for the reverse transcription step in two-step assays of RT-qPCR. Table 2. Primer considerations for the cDNA synthesis step of RT-qPCR. Combining random primers and anchored oligo(dT) primers improves the reverse transcription efficiency and qPCR sensitivity. Primer Options Structure and Function Advantages Oligo(dT)s (or Stretch of thymine residues that anneal to Generation of full length cDNA Only amplify gene with a anchored poly(A) tail of mRNA; anchored from poly(A)-tailed mRNA poly(A) tail oligo(dT)s) oligo(dT)s contain one G, C, or A (the Good to use if little starting Truncated cDNA from anchor) residue at the 3′ end material is available priming internal poly(A) Anchor ensures that the sites*2 oligo(dT) primer binds at the 5′ Bias towards 3′ end* end of the poly(A) tail of mRNA Disadvantages *Minimized if anchored oligo(dT)s are used Random Six to nine bases long, they anneal at Anneal to all RNA (tRNA, rRNA, cDNA is made from all RNAs Primers multiple points along RNA transcript and mRNA) which is not always desirable Good to use for transcripts with and can dilute mRNA signal significant secondary structures, Truncated cDNA or if little starting material is available High cDNA yield Sequence Custom made primers that target specific Specific cDNA pool Synthesis is limited to one Specific Primers mRNA sequence Increased sensitivity gene of interest Use reverse qPCR primer Reverse Transcriptase Enzymes Reverse Transcriptase is the enzyme that makes DNA from RNA. Some enzymes have RNase activity to degrade the RNA strand in the RNA-DNA hybrid after transcription. If an enzyme does not possess RNase activity, an RNaseH may be added for better qPCR efficiency. Commonly used enzymes include Moloney murine leukemia virus reverse transcriptase and Avian myeloblastosis virus reverse transcriptase. For RT-qPCR, it is ideal to choose a reverse transcriptase with high thermal stability, because this allows cDNA synthesis to be performed at higher temperatures, ensuring successful transcription of RNA with high levels of secondary structure, while maintaining their full activity throughout the reaction producing higher cDNA yields. 8/31/2019, 3:29 PM Basic Principles of RT-qPCR | Thermo Fisher Scientific - IN 3 of 4 https://www.thermofisher.com/in/en/home/brands/thermo-scientific/mo... RNase H Activity of Reverse Transcriptase RNase H activity degrades RNA from RNA-DNA duplexes to allow efficient synthesis of double-stranded DNA. However, with long mRNA templates, RNA may be degraded prematurely resulting in truncated cDNA. Hence, it is generally beneficial to minimize RNase H activity when aiming to produce long transcripts for cDNA cloning. In contrast, reverse transcriptases with intrinsic RNase H activity are often favored in qPCR applications because they enhance the melting of RNA-DNA duplex during the first cycles of PCR (Figure 3). Figure 3. RNase H Activity of reverse transcriptases. In qPCR, use a reverse transcriptase with RNAse activity. cDNA Synthesis in a Thermal Cycler Step 1 Step 2 Step 3 Step 4 Predenaturation (Optional) Primer Extension cDNA Synthesis Reaction Termination This step is recommended if the RNA template has a high degree of secondary structure. qPCR in RT-qPCR Primer Design PCR primers for the qPCR step of RT-qPCR should ideally be designed to span an exon-exon junction, with one of the amplification primers potentially spanning the actual exon-intron boundary (Figure 4). This design reduces the risk of false positives from amplification of any contaminating genomic DNA, since the intron-containing genomic DNA sequence would not be amplified. If primers cannot be designed to separate exons or exon-exon boundaries, it is necessary to treat the RNA sample with RNase-free DNase I or dsDNase in order to remove contaminating genomic DNA. Controls for RT-qPCR Click image to enlarge Figure 4. Primer Design for the qPCR step of RT-qPCR. 1) If one primer is designed A minus Reverse Transcription control (-RT control) should be included in all RT-qPCR experiments to test for contaminating DNA (such as genomic DNA or PCR product from a previous run). Such a control contains all the reaction components except for the reverse transcriptase. Reverse transcription should not occur in this control, so if PCR amplification is to span an exon-intron boundary, the possible contaminating genomic DNA is not amplified, because the primer cannot anneal to the template. In contrast, cDNA does not contain any introns, and is efficiently primed and amplified. 2) When primers flank a long (e.g. 1 kb) intron, the amplification cannot occur because the short extension time is sufficient for the short cDNA sequence but not for the longer genomic target. seen, it is most likely derived from contaminating DNA. References 1. Bustin S. (ed) (2004) A-Z of Quantitative PCR. IUL Biotechnology Series, International University Line, La Jolla, California. For Research Use Only. Not for use in diagnostic procedures. India Terms & Conditions Privacy Policy Price & Freight Policy 8/31/2019, 3:29 PM Basic Principles of RT-qPCR | Thermo Fisher Scientific - IN 4 of 4 https://www.thermofisher.com/in/en/home/brands/thermo-scientific/mo... page info 8/31/2019, 3:29 PM