Bernard J. Fisher, Ignacio M. Seropian, Donatas Kraskauskas, Jay N
advertisement

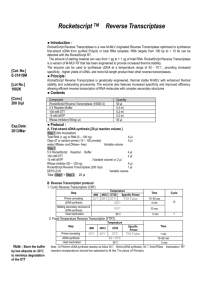
Ascorbic Acid Attenuates Lipopolysaccharide Induced Acute Lung Injury Bernard J. Fisher, Ignacio M. Seropian, Donatas Kraskauskas, Jay N. Thakkar, Norbert F. Voelkel, Alpha A. Fowler III and Ramesh Natarajan ONLINE DATA SUPPLEMENT ADDITIONAL METHODS Reagents and Chemicals LPS from Escherichia coli 0111:B4, AA, DHA, pentobarbital, protease and phosphatase inhibitor cocktails and other chemicals were purchased from SigmaAldrich (St. Louis, MO, USA). The Biopulverizer was purchased from Biospec, Bartlesville, OK. Culture media, serum, NuPAGE™ Novex pre-cast gel system and primers were obtained from GIBCO-Invitrogen (Carlsbad, CA). Sterile tissue culture plasticware was obtained from Corning (Corning, NY). The RNA isolation kit RNeasy™ Mini and QIAshredder™ were obtained from Qiagen (Valencia, CA). High Capacity cDNA Reverse Transcription kit and POWER SYBR Green QPCR Master Mix were obtained from Applied Biosystems Inc. (Foster City, CA). The Renaissance Western Blot Chemiluminescence Reagent Plus was purchased from Perkin Elmer Life Sciences Inc (Boston, MA). Rabbit polyclonal antibodies to lamin B (sc-6216), actin (sc-1616), and NFB p65 (sc-372) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Immobilon membranes were obtained from Millipore (Bedford, MA). Blots were stripped using the Restore™ Western Blot Stripping Buffer (Pierce Biotechnology) as described by the manufacturer.The Vectastain® Elite ABC kit and DAB substrate kit were obtained from Vector Laboratories (Burlingame, CA). Endothelial Cell Culture HMEC-1 cells were cultured in medium MCDB-131 supplemented with 10% FBS, hydrocortisone (1 µg/ml), and epidermal cell growth factor (10 ng/ml) under a 5% CO2 atmosphere at 37ºC. For total RNA preparation and nuclear/cytosolic extracts HMEC-1 were cultured in 35 mm (9.6 cm²) dishes. Western Blot Analysis Proteins were resolved by SDS polyacrylamide gel electrophoresis (4-20%) and electrophoretically transferred to polyvinylidene fluoride membranes (0.2µm pore size). Immunodetection was performed using chemiluminescent detection. Blots were stripped using the Restore™ Western Blot Stripping Buffer as described by the manufacturer. RNA Isolation and Real Time Quantitative PCR (QPCR) Analysis Total RNA was extracted and purified using QIAshredders™ and RNeasy™ columns according to the manufacturer’s specifications (Qiagen). Murine lungs were snap frozen in liquid nitrogen and subsequently powdered with a Biopulverizer (RPI) prior to RNA extraction. Total RNA (1µg) was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit. Complimentary DNA (cDNA) was diluted (1:500) and real time QPCR performed using POWER SYBR Green QPCR Master Mix along with primers (Supplement Table 1, see below). Primers were designed to anneal to sequences on separate exons or to span two exons. Cycling parameters were: 95ºC, 10 min, 40 cycles of 95ºC, 15 sec; 60ºC, 1min. A dissociation profile was generated after each run to verify specificity of amplification. All PCR assays were performed in triplicate. No template controls and no reverse transcriptase controls were included. Beta-actin was used as housekeeping gene against which all the samples were normalized for differences in the amount of total RNA added to each cDNA reaction and for variation in the reverse transcriptase efficiency among the different cDNA reactions. Automated gene expression analysis was performed using the Comparative Quantitation module of MxPro QPCR Software (Stratagene). Supplement Table 1 Name Sequence 5’ to 3’ Product size KC forward KC reverse CAATGAGCTGCGCTGTCAGTGCCTGCAG CTGAACCAAGGGAGCTTCAGGGTC 168bp MIP-2 forward MIP-2 reverse CTGGGGAGAGGGTGAGTTG GCTGTTCTACTCTCCTCGGTG 166bp LIX forward LIX reverse GCGTTGTGTTTGCTTAACCGTAACTCC AGTTTAGCTATGACTTCCACCGTAGGGC 110bp Mpo forward Mpo reverse CTGGATCATGACATCACCTTGACTCC 215bp GATCTGGTTGCGAATGGTGATGTTGTTCC -actin forward -actin reverse MCP-1 forward MCP-1 reverse TCTACGAGGGCTATGCTCTCC TCTTTGATGTCACGCACGATTTC 125bp TTCTGGGCCTGCTGTTCACAG CCAGCCTACTCATTGGGATCATCTTGC 125bp